3MDG
 
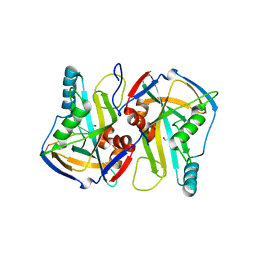 | |
3VGL
 
 | | Crystal structure of a ROK family glucokinase from Streptomyces griseus in complex with glucose and AMPPNP | | 分子名称: | Glucokinase, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, SODIUM ION, ... | | 著者 | Miyazono, K, Tabei, N, Morita, S, Ohnishi, Y, Horinouchi, S, Tanokura, M. | | 登録日 | 2011-08-15 | | 公開日 | 2011-12-07 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Substrate recognition mechanism and substrate-dependent conformational changes of an ROK family glucokinase from Streptomyces griseus
J.Bacteriol., 194, 2012
|
|
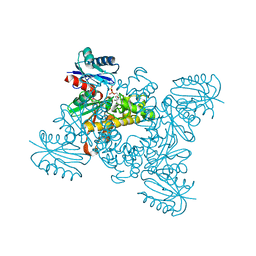
5AWE
 
 | | Crystal structure of a hypothetical protein, TTHA0829 from Thermus thermophilus HB8, composed of cystathionine-beta-synthase (CBS) and aspartate-kinase chorismate-mutase tyrA (ACT) domains | | 分子名称: | Putative acetoin utilization protein, acetoin dehydrogenase | | 著者 | Nakabayashi, M, Shibata, N, Kanagawa, M, Nakagawa, N, Kuramitsu, S, Higuchi, Y. | | 登録日 | 2015-07-03 | | 公開日 | 2016-05-18 | | 最終更新日 | 2020-02-26 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Crystal structure of a hypothetical protein, TTHA0829 from Thermus thermophilus HB8, composed of cystathionine-beta-synthase (CBS) and aspartate-kinase chorismate-mutase tyrA (ACT) domains.
Extremophiles, 20, 2016
|
|
1NYJ
 
 | |
1OHK
 
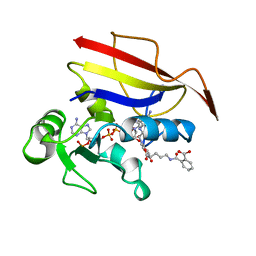 | | HUMAN DIHYDROFOLATE REDUCTASE, ORTHORHOMBIC (P21 21 21) CRYSTAL FORM | | 分子名称: | DIHYDROFOLATE REDUCTASE, N-(4-CARBOXY-4-{4-[(2,4-DIAMINO-PTERIDIN-6-YLMETHYL)-AMINO]-BENZOYLAMINO}-BUTYL)-PHTHALAMIC ACID, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Cody, V, Galitsky, N, Luft, J.R, Pangborn, W. | | 登録日 | 1997-09-17 | | 公開日 | 1998-05-27 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Comparison of two independent crystal structures of human dihydrofolate reductase ternary complexes reduced with nicotinamide adenine dinucleotide phosphate and the very tight-binding inhibitor PT523.
Biochemistry, 36, 1997
|
|
5IRU
 
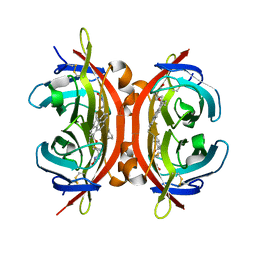 | |
2M5U
 
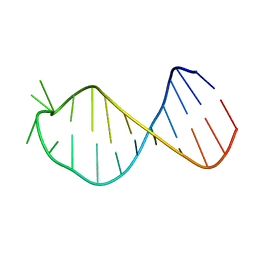 | |
3NZP
 
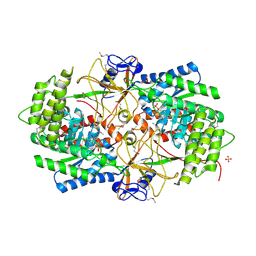 | | Crystal Structure of the Biosynthetic Arginine decarboxylase SpeA from Campylobacter jejuni, Northeast Structural Genomics Consortium Target BR53 | | 分子名称: | Arginine decarboxylase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | 著者 | Forouhar, F, Lew, S, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Belote, R.L, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2010-07-16 | | 公開日 | 2010-09-01 | | 最終更新日 | 2012-02-22 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structures of bacterial biosynthetic arginine decarboxylases.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
5B4T
 
 | | Crystal structure of D-3-hydroxybutyrate dehydrogenase from Alcaligenes faecalis complexed with NAD+ and a substrate D-3-hydroxybutyrate | | 分子名称: | (3R)-3-hydroxybutanoic acid, 3-hydroxybutyrate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | 著者 | Kanazawa, H, Tsunoda, M, Hoque, M.M, Suzuki, K, Yamamoto, T, Takenaka, A. | | 登録日 | 2016-04-19 | | 公開日 | 2016-08-17 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.19 Å) | | 主引用文献 | Structural insights into the catalytic reaction trigger and inhibition of D-3-hydroxybutyrate dehydrogenase
Acta Crystallogr.,Sect.F, 72, 2016
|
|
6J7U
 
 | |
5B4U
 
 | | Crystal structure of D-3-hydroxybutyrate dehydrogenase from Alcaligenes faecalis complexed with NAD+ and an inhibitor malonate | | 分子名称: | 3-hydroxybutyrate dehydrogenase, CHLORIDE ION, MALONIC ACID, ... | | 著者 | Kanazawa, H, Tsunoda, M, Hoque, M.M, Suzuki, K, Yamamoto, T, Takenaka, A. | | 登録日 | 2016-04-19 | | 公開日 | 2016-08-17 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Structural insights into the catalytic reaction trigger and inhibition of D-3-hydroxybutyrate dehydrogenase
Acta Crystallogr.,Sect.F, 72, 2016
|
|
5BSZ
 
 | | X-ray structure of the sugar N-methyltransferase KedS8 from Streptoalloteichus sp ATCC 53650 | | 分子名称: | 1,2-ETHANEDIOL, N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE, ... | | 著者 | Thoden, J.B, Delvaux, N.A, Holden, H.M. | | 登録日 | 2015-06-02 | | 公開日 | 2015-07-29 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Molecular architecture of KedS8, a sugar N-methyltransferase from Streptoalloteichus sp. ATCC 53650.
Protein Sci., 24, 2015
|
|
5BUA
 
 | |
3NSE
 
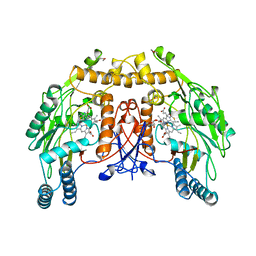 | | BOVINE ENOS, H4B-FREE, SEITU COMPLEX | | 分子名称: | ACETATE ION, ARGININE, CACODYLATE ION, ... | | 著者 | Raman, C.S, Li, H, Martasek, P, Kral, V, Masters, B.S.S, Poulos, T.L. | | 登録日 | 1998-09-23 | | 公開日 | 1999-05-18 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal structure of constitutive endothelial nitric oxide synthase: a paradigm for pterin function involving a novel metal center.
Cell(Cambridge,Mass.), 95, 1998
|
|
2ZOY
 
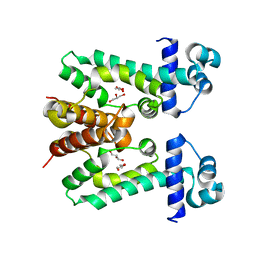 | | The multi-drug binding transcriptional repressor CgmR (CGL2612 protein) from C.glutamicum | | 分子名称: | GLYCEROL, Transcriptional regulator | | 著者 | Itou, H, Yao, M, Watanabe, N, Tanaka, I. | | 登録日 | 2008-06-20 | | 公開日 | 2008-07-08 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | The CGL2612 protein from Corynebacterium glutamicum is a drug resistance-related transcriptional repressor: structural and functional analysis of a newly identified transcription factor from genomic DNA analysis
J.Biol.Chem., 280, 2005
|
|
3O48
 
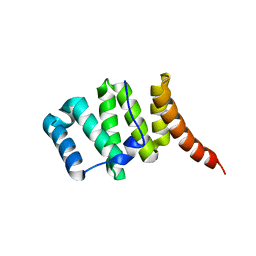 | | Crystal structure of fission protein Fis1 from Saccharomyces cerevisiae | | 分子名称: | Mitochondria fission 1 protein | | 著者 | Tooley, J.E, Khangulov, V, Heroux, A, Bosch, J, Hill, R.B. | | 登録日 | 2010-07-26 | | 公開日 | 2011-08-10 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | The 1.75 Angstrom resolution structure of fission protein Fis1 from Saccharomyces cerevisiae reveals elusive interactions of the autoinhibitory domain
Acta Crystallogr.,Sect.F, 67, 2011
|
|
3O83
 
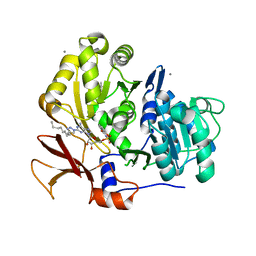 | | Structure of BasE N-terminal domain from Acinetobacter baumannii bound to 2-(4-n-dodecyl-1,2,3-triazol-1-yl)-5'-O-[N-(2-hydroxybenzoyl)sulfamoyl]adenosine | | 分子名称: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(4-dodecyl-1H-1,2,3-triazol-1-yl)-5'-O-{[(2-hydroxyphenyl)carbonyl]sulfamoyl}adenosine, ... | | 著者 | Drake, E.J, Duckworth, B.P, Neres, J, Aldrich, C.C, Gulick, A.M. | | 登録日 | 2010-08-02 | | 公開日 | 2010-10-06 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Biochemical and structural characterization of bisubstrate inhibitors of BasE, the self-standing nonribosomal peptide synthetase adenylate-forming enzyme of acinetobactin synthesis.
Biochemistry, 49, 2010
|
|
5J6F
 
 | | Crystal structure of DAH7PS-CM complex from Geobacillus sp. with prephenate | | 分子名称: | 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase, chorismate mutase-isozyme 3, MANGANESE (II) ION, ... | | 著者 | Nazmi, A.R, Othman, M, Lang, E.J.M, Bai, Y, Allison, T.M, Panjkar, S, Arcus, V.L, Parker, E.J. | | 登録日 | 2016-04-04 | | 公開日 | 2016-09-07 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Interdomain Conformational Changes Provide Allosteric Regulation en Route to Chorismate.
J. Biol. Chem., 291, 2016
|
|
1WOS
 
 | | Crystal Structure of T-protein of the Glycine Cleavage System | | 分子名称: | Aminomethyltransferase | | 著者 | Lee, H.H, Kim, D.J, Ahn, H.J, Ha, J.Y, Suh, S.W. | | 登録日 | 2004-08-24 | | 公開日 | 2004-09-07 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.84 Å) | | 主引用文献 | Crystal Structure of T-protein of the Glycine Cleavage System: Cofactor binding, insights into H-protein recognition, and molecular basis for understanding nonketotic hyperglycinemia
J.Biol.Chem., 279, 2004
|
|
3O84
 
 | | Structure of BasE N-terminal domain from Acinetobacter baumannii bound to 6-phenyl-1-(pyridin-4-ylmethyl)-1H-pyrazolo[3,4-b]pyridine-4-carboxylic acid. | | 分子名称: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 6-phenyl-1-(pyridin-4-ylmethyl)-1H-pyrazolo[3,4-b]pyridine-4-carboxylic acid, ... | | 著者 | Drake, E.J, Duckworth, B.P, Neres, J, Aldrich, C.C, Gulick, A.M. | | 登録日 | 2010-08-02 | | 公開日 | 2010-10-06 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Biochemical and structural characterization of bisubstrate inhibitors of BasE, the self-standing nonribosomal peptide synthetase adenylate-forming enzyme of acinetobactin synthesis.
Biochemistry, 49, 2010
|
|
3O82
 
 | | Structure of BasE N-terminal domain from Acinetobacter baumannii bound to 5'-O-[N-(2,3-dihydroxybenzoyl)sulfamoyl] adenosine | | 分子名称: | 5'-O-{[(2,3-dihydroxyphenyl)carbonyl]sulfamoyl}adenosine, CALCIUM ION, Peptide arylation enzyme | | 著者 | Drake, E.J, Duckworth, B.P, Neres, J, Aldrich, C.C, Gulick, A.M. | | 登録日 | 2010-08-02 | | 公開日 | 2010-10-06 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Biochemical and structural characterization of bisubstrate inhibitors of BasE, the self-standing nonribosomal peptide synthetase adenylate-forming enzyme of acinetobactin synthesis.
Biochemistry, 49, 2010
|
|
6V38
 
 | | Cryo-EM structure of Ca2+-bound hsSlo1 channel | | 分子名称: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, CALCIUM ION, CHOLESTEROL, ... | | 著者 | Tao, X, MacKinnon, R. | | 登録日 | 2019-11-25 | | 公開日 | 2019-12-25 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Molecular structures of the human Slo1 K + channel in complex with beta 4.
Elife, 8, 2019
|
|
6JKU
 
 | | Crystal structure of N-acetylglucosamine-6-phosphate deacetylase from Pasteurella Multocida | | 分子名称: | 1,2-ETHANEDIOL, GLYCEROL, N-acetylglucosamine-6-phosphate deacetylase, ... | | 著者 | Manjunath, L, Bose, S, Subramanian, R. | | 登録日 | 2019-03-01 | | 公開日 | 2020-03-04 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Quaternary variations in the structural assembly of N-acetylglucosamine-6-phosphate deacetylase from Pasteurella multocida.
Proteins, 2020
|
|
5O4V
 
 | |
5JYF
 
 | |
