8E4V
 
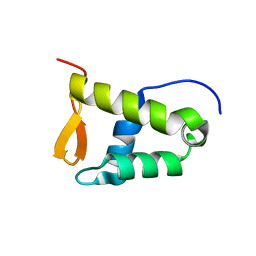 | |
5JNO
 
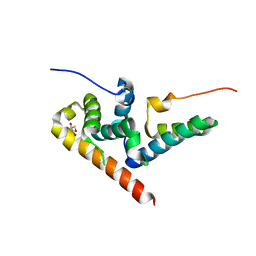 | |
5VA6
 
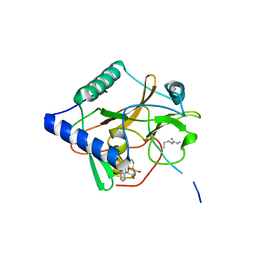 | | CRYSTAL STRUCTURE OF ATXR5 IN COMPLEX WITH HISTONE H3.1 MONO-METHYLATED ON R26 | | 分子名称: | Histone H3.1, Probable Histone-lysine N-methyltransferase ATXR5, S-ADENOSYL-L-HOMOCYSTEINE | | 著者 | Bergamin, E, Sarvan, S, Malette, J, Eram, M, Yeung, S, Mongeon, V, Joshi, M, Brunzelle, J.S, Michaels, S.D, Blais, A, Vedadi, M, Couture, J.-F. | | 登録日 | 2017-03-24 | | 公開日 | 2017-04-19 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Molecular basis for the methylation specificity of ATXR5 for histone H3.
Nucleic Acids Res., 45, 2017
|
|
5VAH
 
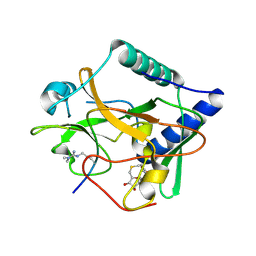 | | Crystal structure of ATXR5 SET domain in complex with histone H3 di-methylated on R26 | | 分子名称: | Histone H3.2, Probable Histone-lysine N-methyltransferase ATXR5, S-ADENOSYL-L-HOMOCYSTEINE | | 著者 | Bergamin, E, Sarvan, S, Malette, J, Eram, M, Yeung, S, Mongeon, V, Joshi, M, Brunzelle, J.S, Michaels, S.D, Blais, A, Vedadi, M, Couture, J.-F. | | 登録日 | 2017-03-26 | | 公開日 | 2017-04-05 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Molecular basis for the methylation specificity of ATXR5 for histone H3.
Nucleic Acids Res., 45, 2017
|
|
5VBC
 
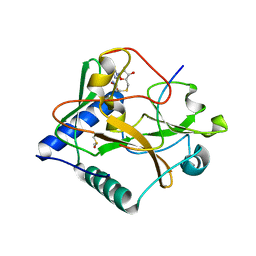 | |
5VAB
 
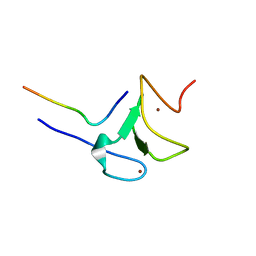 | | Crystal structure of ATXR5 PHD domain in complex with histone H3 | | 分子名称: | ATXR5 PHD domain, Histone H3 peptide, ZINC ION | | 著者 | Bergamin, E, Sarvan, S, Malette, J, Eram, M, Yeung, S, Mongeon, V, Joshi, M, Brunzelle, J.S, Michaels, S.D, Blais, A, Vedadi, M, Couture, J.-F. | | 登録日 | 2017-03-24 | | 公開日 | 2017-04-19 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.702 Å) | | 主引用文献 | Molecular basis for the methylation specificity of ATXR5 for histone H3.
Nucleic Acids Res., 45, 2017
|
|
5W3V
 
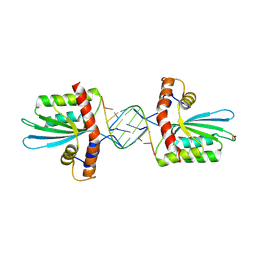 | | Crystal Structure of macaque APOBEC3H in complex with RNA | | 分子名称: | Apobec3H, RNA (5'-R(P*AP*AP*CP*CP*CP*CP*GP*GP*GP*C)-3'), RNA (5'-R(P*AP*AP*CP*CP*CP*GP*GP*GP*GP*A)-3'), ... | | 著者 | Bohn, J.A, Thummar, K, York, A, Raymond, A, Brown, W.C, Bieniasz, P.D, Hatziioannou, T, Smith, J.L. | | 登録日 | 2017-06-08 | | 公開日 | 2017-10-25 | | 最終更新日 | 2017-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.243 Å) | | 主引用文献 | APOBEC3H structure reveals an unusual mechanism of interaction with duplex RNA.
Nat Commun, 8, 2017
|
|
2XI8
 
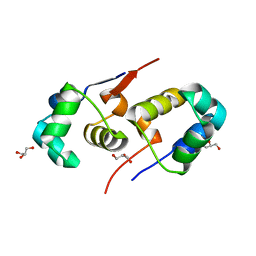 | | High resolution structure of native CylR2 | | 分子名称: | GLYCEROL, PUTATIVE TRANSCRIPTION REGULATOR | | 著者 | Gruene, T, Cho, M.-K, Karyagina, I, Kim, H.-Y, Grosse, C, Giller, K, Zweckstetter, M, Becker, S. | | 登録日 | 2010-06-28 | | 公開日 | 2011-02-09 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.21 Å) | | 主引用文献 | Integrated Analysis of the Conformation of a Protein-Linked Spin Label by Crystallography, Epr and NMR Spectroscopy.
J.Biomol.NMR, 49, 2011
|
|
1TBA
 
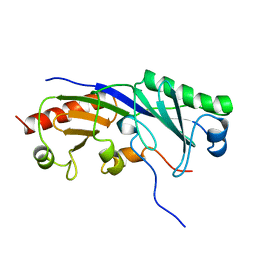 | | SOLUTION STRUCTURE OF A TBP-TAFII230 COMPLEX: PROTEIN MIMICRY OF THE MINOR GROOVE SURFACE OF THE TATA BOX UNWOUND BY TBP, NMR, 25 STRUCTURES | | 分子名称: | TRANSCRIPTION INITIATION FACTOR IID 230K CHAIN, TRANSCRIPTION INITIATION FACTOR TFIID | | 著者 | Liu, D, Ishima, R, Tong, K.I, Bagby, S, Kokubo, T, Muhandiram, D.R, Kay, L.E, Nakatani, Y, Ikura, M. | | 登録日 | 1998-08-16 | | 公開日 | 1999-08-16 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of a TBP-TAF(II)230 complex: protein mimicry of the minor groove surface of the TATA box unwound by TBP.
Cell(Cambridge,Mass.), 94, 1998
|
|
3GA8
 
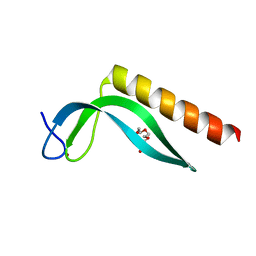 | | Structure of the N-terminal domain of the E. coli protein MqsA (YgiT/b3021) | | 分子名称: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, HTH-type transcriptional regulator MqsA (YgiT/B3021), ZINC ION | | 著者 | Brown, B.L, Arruda, J.M, Peti, W, Page, R. | | 登録日 | 2009-02-16 | | 公開日 | 2010-01-12 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Three dimensional structure of the MqsR:MqsA complex: a novel TA pair comprised of a toxin homologous to RelE and an antitoxin with unique properties.
Plos Pathog., 5, 2009
|
|
2F2E
 
 | | Crystal Structure of PA1607, a Putative Transcription Factor | | 分子名称: | PA1607, SULFATE ION, alpha-D-glucopyranose | | 著者 | Sieminska, E.A, Xu, X, Zheng, H, Lunin, V, Cuff, M, Joachimiak, A, Edwards, A, Savchenko, A, Sanders, D.A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2005-11-16 | | 公開日 | 2006-03-14 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | The X-ray crystal structure of PA1607 from Pseudomonas aureginosa at 1.9 A resolution--a putative transcription factor.
Protein Sci., 16, 2007
|
|
1HHW
 
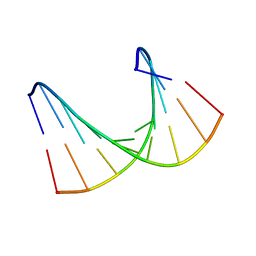 | | Solution structure of LNA1:RNA hybrid | | 分子名称: | 5- D(*CP*TP*GP*AP*+TLNP*AP*TP*GP*C) -3, 5- R(*GP*CP*AP*UP*AP*UP*CP*AP*G) -3 | | 著者 | Petersen, M, Bondensgaard, K, Wengel, J, Jacobsen, J.P. | | 登録日 | 2000-12-29 | | 公開日 | 2002-05-30 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Locked Nucleic Acid (Lna) Recognition of RNA: NMR Solution Structures of Lna:RNA Hybrids
J.Am.Chem.Soc., 124, 2002
|
|
1HMV
 
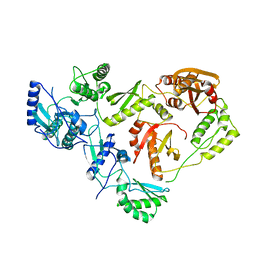 | | THE STRUCTURE OF UNLIGANDED REVERSE TRANSCRIPTASE FROM THE HUMAN IMMUNODEFICIENCY VIRUS TYPE 1 | | 分子名称: | HIV-1 REVERSE TRANSCRIPTASE (SUBUNIT P51), HIV-1 REVERSE TRANSCRIPTASE (SUBUNIT P66), MAGNESIUM ION | | 著者 | Rodgers, D.W, Gamblin, S.J, Harris, B.A, Ray, S, Culp, J.S, Hellmig, B, Woolf, D.J, Debouck, C, Harrison, S.C. | | 登録日 | 1994-12-15 | | 公開日 | 1995-03-31 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | The structure of unliganded reverse transcriptase from the human immunodeficiency virus type 1.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
1HVU
 
 | |
2QM4
 
 | | Crystal structure of human XLF/Cernunnos, a non-homologous end-joining factor | | 分子名称: | Non-homologous end-joining factor 1 | | 著者 | Li, Y, Chirgadze, D.Y, Sibanda, B.L, Bolanos-Garcia, V.M, Davies, O.R, Blundell, T.L. | | 登録日 | 2007-07-14 | | 公開日 | 2007-12-11 | | 最終更新日 | 2017-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structure of human XLF/Cernunnos reveals unexpected differences from XRCC4 with implications for NHEJ.
Embo J., 27, 2008
|
|
1MXK
 
 | | NMR Structure of HO2-Co(III)bleomycin A(2) Bound to d(GGAAGCTTCC)(2) | | 分子名称: | 5'-D(*GP*GP*AP*AP*GP*CP*TP*TP*CP*C)-3', BLEOMYCIN A2, COBALT (III) ION, ... | | 著者 | Zhao, C, Xia, C, Mao, Q, Forsterling, H, DeRose, E, Antholine, W.E, Subczynski, W.K, Petering, D.H. | | 登録日 | 2002-10-02 | | 公開日 | 2002-10-16 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structures of HO(2)-Co(III)bleomycin A(2) Bound to d(GAGCTC)(2) and d(GGAAGCTTCC)(2): Structure-Reactivity Relationships of Co and Fe Bleomycins
J.Inorg.Biochem., 91, 2002
|
|
7VII
 
 | |
1MTG
 
 | | NMR Structure of HO2-Co(III)bleomycin A(2) bound to d(GAGCTC)(2) | | 分子名称: | 5'-D(*GP*AP*GP*CP*TP*C)-3', BLEOMYCIN A2, COBALT (III) ION, ... | | 著者 | Zhao, C, Xia, C, Mao, Q, Forsterling, H, DeRose, E, Antholine, W.E, Subczynski, W.K, Petering, D.H. | | 登録日 | 2002-09-20 | | 公開日 | 2002-10-16 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structures of HO(2)-Co(III)bleomycin A(2) Bound to d(GAGCTC)(2) and d(GGAAGCTTCC)(2): Structure-Reactivity Relationships of Co and Fe Bleomycins
J.Inorg.Biochem., 91, 2002
|
|
7VIK
 
 | |
2YDK
 
 | | Discovery of Checkpoint Kinase Inhibitor AZD7762 by Structure Based Design and Optimization of Thiophene Carboxamide Ureas | | 分子名称: | 2-(CARBAMOYLAMINO)-5-PHENYL-N-[(3S)-PIPERIDIN-3-YL]THIOPHENE-3-CARBOXAMIDE, GLYCEROL, SERINE/THREONINE-PROTEIN KINASE CHK1, ... | | 著者 | Read, J.A, Breed, J, Haye, H, McCall, E, Rowsell, S, Vallentine, A, White, A. | | 登録日 | 2011-03-22 | | 公開日 | 2012-04-04 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Discovery of Checkpoint Kinase Inhibitor (S)-5-(3-Fluorophenyl)-N-(Piperidin-3-Yl)-3-Ureidothiophene-2-Carboxamide (Azd7762) by Structure-Based Design and Optimization of Thiophenecarboxamide Ureas.
J.Med.Chem., 55, 2012
|
|
4BUL
 
 | | Novel hydroxyl tricyclics (e.g. GSK966587) as potent inhibitors of bacterial type IIA topoisomerases | | 分子名称: | (S)-4-((4-(((2,3-dihydro-[1,4]dioxino[2,3-c]pyridin-7-yl)methyl)amino)piperidin-1-yl)methyl)-3-fluoro-4-hydroxy-4H-pyrrolo[3,2,1-de][1,5]naphthyridin-7(5H)-one, 5'-D(*AP*GP*CP*CP*GP*TP*AP*GP*GP*TP*AP*CP*AP*CP *CP*GP*CP*AP*C)-3', 5'-D(*TP*GP*TP*GP*CP*GP*GP*TP*GP*TP*AP*CP*CP*TP *AP*CP*GP*GP*CP*T)-3', ... | | 著者 | Miles, T.J, Hennessy, A.J, Bax, B, Brooks, G, Brown, B.S, Brown, P, Cailleau, N, Chen, D, Dabbs, S, Davies, D.T, Esken, J.M, Giordano, I, Hoover, J.L, Huang, J, Jones, G.E, Sukmar, S.K.K, Spitzfaden, C, Markwell, R.E, Minthorn, E.A, Rittenhouse, S, Gwynn, M.N, Pearson, N.D. | | 登録日 | 2013-06-20 | | 公開日 | 2013-08-07 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Novel Hydroxyl Tricyclics (E.G., Gsk966587) as Potent Inhibitors of Bacterial Type Iia Topoisomerases.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
2LSN
 
 | |
1IPI
 
 | |
2YDI
 
 | | Discovery of Checkpoint Kinase Inhibitor AZD7762 by Structure Based Design and Optimization of Thiophene Carboxamide Ureas | | 分子名称: | 5-[4-(2-DIMETHYLAMINOETHYLOXY)PHENYL]-2-UREIDO-THIOPHENE-3-CARBOXAMIDE, SERINE/THREONINE-PROTEIN KINASE CHK1, SULFATE ION | | 著者 | Read, J.A, Breed, J, Haye, H, McCall, E, Rowsell, S, Vallentine, A, White, A. | | 登録日 | 2011-03-21 | | 公開日 | 2012-04-04 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Discovery of Checkpoint Kinase Inhibitor (S)-5-(3-Fluorophenyl)-N-(Piperidin-3-Yl)-3-Ureidothiophene-2-Carboxamide (Azd7762) by Structure-Based Design and Optimization of Thiophenecarboxamide Ureas.
J.Med.Chem., 55, 2012
|
|
7W8D
 
 | | The structure of Deinococcus radiodurans RuvC | | 分子名称: | Crossover junction endodeoxyribonuclease RuvC, MAGNESIUM ION | | 著者 | Cheng, K. | | 登録日 | 2021-12-07 | | 公開日 | 2022-08-17 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.75016141 Å) | | 主引用文献 | Biochemical and Structural Study of RuvC and YqgF from Deinococcus radiodurans.
Mbio, 13, 2022
|
|
