4ZGN
 
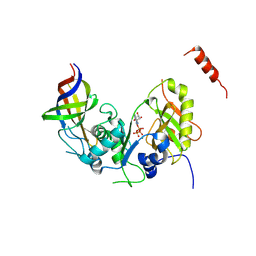 | | Structure Cdc123 complexed with the C-terminal domain of eIF2gamma | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Cell division cycle protein 123, Eukaryotic translation initiation factor 2 subunit gamma, ... | | 著者 | Panvert, M, Dubiez, E, Arnold, L, Perez, J, Seufert, W, Mechulam, Y, Schmitt, E. | | 登録日 | 2015-04-23 | | 公開日 | 2015-09-30 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Cdc123, a Cell Cycle Regulator Needed for eIF2 Assembly, Is an ATP-Grasp Protein with Unique Features.
Structure, 23, 2015
|
|
5E2U
 
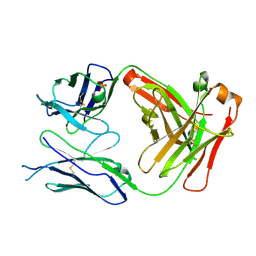 | |
6DQW
 
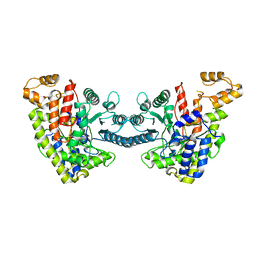 | |
6DID
 
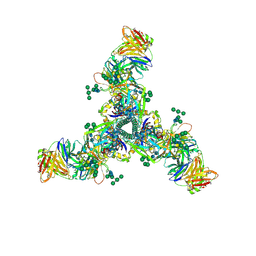 | | HIV Env BG505 SOSIP with polyclonal Fabs from immunized rabbit #3417 post-boost#1 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp160, ... | | 著者 | Turner, H.L, Cottrell, C.A, Oyen, D, Wilson, I.A, Ward, A.B. | | 登録日 | 2018-05-23 | | 公開日 | 2018-09-05 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (4.71 Å) | | 主引用文献 | Electron-Microscopy-Based Epitope Mapping Defines Specificities of Polyclonal Antibodies Elicited during HIV-1 BG505 Envelope Trimer Immunization.
Immunity, 49, 2018
|
|
2ZBY
 
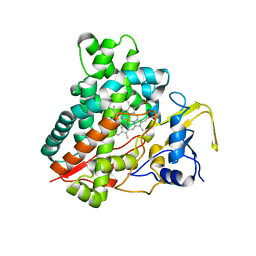 | | Crystal structure of vitamin D hydroxylase cytochrome P450 105A1 (R84A mutant) | | 分子名称: | Cytochrome P450-SU1, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Sugimoto, H, Shinkyo, R, Hayashi, K, Yoneda, S, Yamada, M, Kamakura, M, Ikushiro, S, Shiro, Y, Sakaki, T. | | 登録日 | 2007-10-30 | | 公開日 | 2008-04-08 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystal Structure of CYP105A1 (P450SU-1) in Complex with 1alpha,25-Dihydroxyvitamin D3
Biochemistry, 47, 2008
|
|
5E0C
 
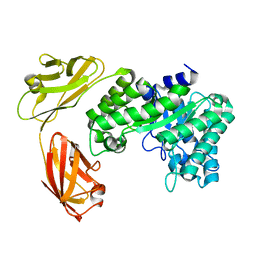 | |
6DKM
 
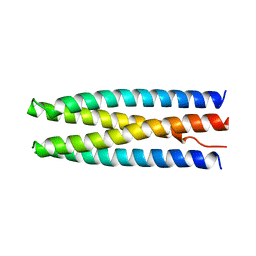 | | DHD131 | | 分子名称: | DHD131_A, DHD131_B | | 著者 | Bick, M.J, Chen, Z, Baker, D. | | 登録日 | 2018-05-29 | | 公開日 | 2018-12-19 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.38 Å) | | 主引用文献 | Programmable design of orthogonal protein heterodimers.
Nature, 565, 2019
|
|
6DKS
 
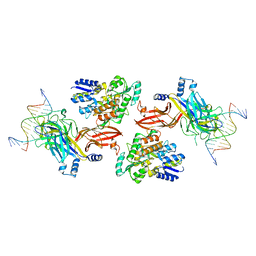 | | Structure of the Rbpj-SHARP-DNA Repressor Complex | | 分子名称: | DNA (5'-D(*AP*AP*TP*CP*TP*TP*TP*CP*CP*CP*AP*CP*AP*GP*T)-3'), DNA (5'-D(*TP*TP*AP*CP*TP*GP*TP*GP*GP*GP*AP*AP*AP*GP*A)-3'), Maltose/maltodextrin-binding periplasmic protein, ... | | 著者 | Kovall, R.A, VanderWielen, B.D, Yuan, Z. | | 登録日 | 2018-05-30 | | 公開日 | 2019-01-02 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.78 Å) | | 主引用文献 | Structural and Functional Studies of the RBPJ-SHARP Complex Reveal a Conserved Corepressor Binding Site.
Cell Rep, 26, 2019
|
|
4F0S
 
 | | Crystal structure of an adenosine deaminase homolog from Chromobacterium violaceum (target NYSGRC-019589) with bound inosine. | | 分子名称: | 5-methylthioadenosine/S-adenosylhomocysteine deaminase, CHLORIDE ION, INOSINE, ... | | 著者 | Kim, J, Vetting, M.W, Sauder, J.M, Burley, S.K, Raushel, F.M, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2012-05-04 | | 公開日 | 2012-06-06 | | 最終更新日 | 2021-02-10 | | 実験手法 | X-RAY DIFFRACTION (1.851 Å) | | 主引用文献 | Crystal structure of an adenosine deaminase homolog from Chromobacterium violaceum (target NYSGRC-019589) with bound inosine.
To be Published
|
|
5E5W
 
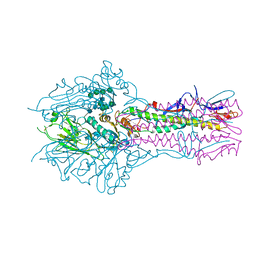 | | Hemagglutinin-esterase-fusion mutant structure of influenza D virus | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin-esterase, ... | | 著者 | Song, H, Qi, J, Shi, Y, Gao, G.F. | | 登録日 | 2015-10-09 | | 公開日 | 2016-03-23 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | An Open Receptor-Binding Cavity of Hemagglutinin-Esterase-Fusion Glycoprotein from Newly-Identified Influenza D Virus: Basis for Its Broad Cell Tropism
PLoS Pathog., 12, 2016
|
|
5E78
 
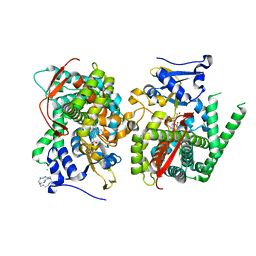 | | Crystal structure of P450 BM3 heme domain variant complexed with Co(III)Sep | | 分子名称: | 1,3,6,8,10,13,16,19-octaazabicyclo[6.6.6]icosane, Bifunctional P-450/NADPH-P450 reductase, CHLORIDE ION, ... | | 著者 | Panneerselvm, S, Shehzad, A, Bocola, M, Mueller-Dieckmann, J, Schwaneberg, U. | | 登録日 | 2015-10-12 | | 公開日 | 2016-09-28 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystallographic insights into a cobalt (III) sepulchrate based alternative cofactor system of P450 BM3 monooxygenase.
Biochim. Biophys. Acta, 1866, 2018
|
|
1YW6
 
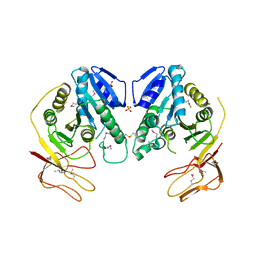 | | Crystal Structure of Succinylglutamate Desuccinylase from Escherichia coli, Northeast Structural Genomics Target ET72. | | 分子名称: | SULFATE ION, Succinylglutamate desuccinylase | | 著者 | Forouhar, F, Yong, W, Kuzin, A.P, Ciano, M, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2005-02-17 | | 公開日 | 2005-03-08 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Crystal Structure of Succinylglutamate Desuccinylase from Escherichia coli, Northeast Structural Genomics Target ET72.
To be Published
|
|
4OWR
 
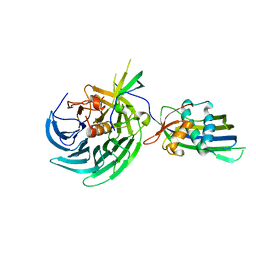 | | Vesiculoviral matrix (M) protein occupies nucleic acid binding site at nucleoporin pair Rae1-Nup98 | | 分子名称: | Matrix protein, Nuclear pore complex protein Nup98-Nup96, mRNA export factor | | 著者 | Ren, Y, Quan, B, Seo, H.S, Blobel, G. | | 登録日 | 2014-02-03 | | 公開日 | 2014-06-25 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (3.15 Å) | | 主引用文献 | Vesiculoviral matrix (M) protein occupies nucleic acid binding site at nucleoporin pair (Rae1 Nup98).
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5DKX
 
 | | Crystal structure of glucosidase II alpha subunit (Tris-bound from) | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Alpha glucosidase-like protein, CHLORIDE ION | | 著者 | Satoh, T, Toshimori, T, Yan, G, Yamaguchi, T, Kato, K. | | 登録日 | 2015-09-04 | | 公開日 | 2016-01-27 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structural basis for two-step glucose trimming by glucosidase II involved in ER glycoprotein quality control.
Sci Rep, 6, 2016
|
|
5DM9
 
 | |
5DN4
 
 | |
5E93
 
 | | Crystal Structure of Trypanosoma cruzi Dihydroorotate Dehydrogenase in Complex with Neq0071 | | 分子名称: | 1,2-ETHANEDIOL, 5-[(E)-3-(furan-2-yl)prop-2-enylidene]-1,3-diazinane-2,4,6-trione, CACODYLATE ION, ... | | 著者 | Rocha, J.R, Inaoka, D.K, Cheleski, J, Shiba, T, Harada, S, Montanari, C.A, Kita, K. | | 登録日 | 2015-10-14 | | 公開日 | 2016-10-19 | | 最終更新日 | 2020-02-19 | | 実験手法 | X-RAY DIFFRACTION (1.41 Å) | | 主引用文献 | Exploring Trypanosoma cruzi Dihydroorotate Dehydrogenase Active Site Plasticity for the Discovery of Potent and Selective Inhibitors with Trypanocidal Activity
To be Published
|
|
6DDE
 
 | | Mu Opioid Receptor-Gi Protein Complex | | 分子名称: | DAMGO, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Koehl, A, Hu, H, Maeda, S, Manglik, A, Zhang, Y, Kobilka, B.K, Skiniotis, G, Weis, W.I. | | 登録日 | 2018-05-10 | | 公開日 | 2018-06-13 | | 最終更新日 | 2023-11-15 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structure of the mu-opioid receptor-Giprotein complex.
Nature, 558, 2018
|
|
5EAE
 
 | | Saccharomyces cerevisiae CYP51 complexed with the plant pathogen inhibitor R-desthio-prothioconazole | | 分子名称: | (2~{R})-2-(1-chloranylcyclopropyl)-1-(2-chlorophenyl)-3-(1,2,4-triazol-1-yl)propan-2-ol, Lanosterol 14-alpha demethylase, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Tyndall, J.D.A, Sabherwal, M, Sagatova, A.A, Keniya, M.V, Wilson, R.K, Woods, M.V, Monk, B.C. | | 登録日 | 2015-10-16 | | 公開日 | 2016-02-10 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.11 Å) | | 主引用文献 | Structural and Functional Elucidation of Yeast Lanosterol 14 alpha-Demethylase in Complex with Agrochemical Antifungals.
PLoS ONE, 11, 2016
|
|
5EB8
 
 | |
5EC2
 
 | | KcsA with V76ester+G77dA mutations | | 分子名称: | Antibody Fab Fragment Light Chain, DIACYL GLYCEROL, NONAN-1-OL, ... | | 著者 | Matulef, K, Valiyaveetil, F.I. | | 登録日 | 2015-10-20 | | 公開日 | 2016-04-20 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Individual Ion Binding Sites in the K(+) Channel Play Distinct Roles in C-type Inactivation and in Recovery from Inactivation.
Structure, 24, 2016
|
|
6DX7
 
 | |
6D6X
 
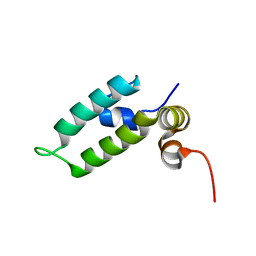 | | HSP40 co-chaperone Sis1 J-domain | | 分子名称: | Type II HSP40 co-chaperone | | 著者 | Pinheiro, G.M.S, Amorim, G.C, Iqbal, A, Ramos, C.H.I, Almeida, F.C.L. | | 登録日 | 2018-04-23 | | 公開日 | 2019-05-01 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution NMR investigation on the structure and function of the isolated J-domain from Sis1: Evidence of transient inter-domain interactions in the full-length protein.
Arch.Biochem.Biophys., 669, 2019
|
|
6DM9
 
 | | DHD15_extended | | 分子名称: | DHD15_extended_A, DHD15_extended_B, SULFATE ION | | 著者 | Bick, M.J, Chen, Z, Baker, D. | | 登録日 | 2018-06-04 | | 公開日 | 2018-12-19 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Programmable design of orthogonal protein heterodimers.
Nature, 565, 2019
|
|
5DZE
 
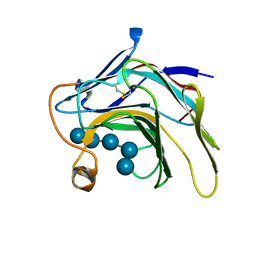 | | Crystal Structure of the catalytic nucleophile mutant of VvEG16 in complex with cellotetraose | | 分子名称: | beta-D-glucopyranose, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose, endo-glucanase | | 著者 | McGregor, N.G.S, Tung, C.C, Van Petegem, F, Brumer, H. | | 登録日 | 2015-09-25 | | 公開日 | 2016-09-21 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (0.97 Å) | | 主引用文献 | Crystallographic insight into the evolutionary origins of xyloglucan endotransglycosylases and endohydrolases.
Plant J., 89, 2017
|
|
