8BSJ
 
 | | Giardia Ribosome in PRE-T Classical State (C) | | 分子名称: | 40S ribosomal protein S21, 40S ribosomal protein S25, 40S ribosomal protein S26, ... | | 著者 | Majumdar, S, Emmerich, A.G, Sanyal, S. | | 登録日 | 2022-11-25 | | 公開日 | 2023-03-22 | | 最終更新日 | 2023-05-03 | | 実験手法 | ELECTRON MICROSCOPY (6.49 Å) | | 主引用文献 | Insights into translocation mechanism and ribosome evolution from cryo-EM structures of translocation intermediates of Giardia intestinalis.
Nucleic Acids Res., 51, 2023
|
|
8BQD
 
 | | Yeast 80S ribosome in complex with Map1 (conformation 1) | | 分子名称: | 18S rRNA, 25S rRNA, 40S ribosomal protein S0-A, ... | | 著者 | Knorr, A.G, Mackens-Kiani, T, Musial, J, Berninghausen, O, Becker, T, Beatrix, B, Beckmann, R. | | 登録日 | 2022-11-21 | | 公開日 | 2023-03-22 | | 最終更新日 | 2023-05-03 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | The dynamic architecture of Map1- and NatB-ribosome complexes coordinates the sequential modifications of nascent polypeptide chains.
Plos Biol., 21, 2023
|
|
7XNY
 
 | |
7XNX
 
 | |
8F6P
 
 | | Rat Cardiac Sodium Channel with Ranolazine Bound | | 分子名称: | (R)-ranolazine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Lenaeus, M.J, Tonggu, L. | | 登録日 | 2022-11-16 | | 公開日 | 2023-04-12 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structural Basis for Inhibition of the Cardiac Sodium Channel by the Atypical Antiarrhythmic Drug Ranolazine
To Be Published
|
|
7Z3N
 
 | | Cryo-EM structure of the ribosome-associated RAC complex on the 80S ribosome - RAC-1 conformation | | 分子名称: | 18S rRNA, 26S rRNA, 40S ribosomal protein S0, ... | | 著者 | Kisonaite, M, Wild, K, Sinning, I. | | 登録日 | 2022-03-02 | | 公開日 | 2023-04-12 | | 最終更新日 | 2023-05-31 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structural inventory of cotranslational protein folding by the eukaryotic RAC complex.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7Z3O
 
 | | Cryo-EM structure of the ribosome-associated RAC complex on the 80S ribosome - RAC-2 conformation | | 分子名称: | 18S rRNA, 26S rRNA, 40S ribosomal protein S0, ... | | 著者 | Kisonaite, M, Wild, K, Sinning, I. | | 登録日 | 2022-03-02 | | 公開日 | 2023-04-12 | | 最終更新日 | 2023-05-31 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structural inventory of cotranslational protein folding by the eukaryotic RAC complex.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8BVG
 
 | |
7Q6B
 
 | | mRubyFT/S148I, a mutant of blue-to-red fluorescent timer in its blue state | | 分子名称: | mRubyFT S148I, a mutant of blue-to-red fluorescent timer | | 著者 | Boyko, K.M, Nikolaeva, A.Y, Vlaskina, A.V, Dorovatovskii, P.V, Khrenova, M.G, Subach, O.M, Popov, V.O, Subach, F.M. | | 登録日 | 2021-11-06 | | 公開日 | 2023-04-12 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Combined Structural and Computational Study of the mRubyFT Fluorescent Timer Locked in Its Blue Form.
Int J Mol Sci, 24, 2023
|
|
8S9Y
 
 | |
8FCK
 
 | | Structure of the vertebrate augmin complex | | 分子名称: | HAUS augmin like complex subunit 2 L homeolog, Green fluorescent protein chimera, HAUS augmin like complex subunit 4 L homeolog, ... | | 著者 | Travis, S.M, Huang, W, Zhang, R, Petry, S. | | 登録日 | 2022-12-01 | | 公開日 | 2023-04-19 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (6.88 Å) | | 主引用文献 | Integrated model of the vertebrate augmin complex.
Nat Commun, 14, 2023
|
|
8GLP
 
 | | mRNA decoding in human is kinetically and structurally distinct from bacteria (Consensus LSU focused refined structure) | | 分子名称: | 1,4-DIAMINOBUTANE, 18S rRNA, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | 著者 | Holm, M, Natchiar, K.S, Rundlet, E.J, Myasnikov, A.G, Watson, Z.L, Altman, R.B, Blanchard, S.C. | | 登録日 | 2023-03-22 | | 公開日 | 2023-04-19 | | 最終更新日 | 2023-11-15 | | 実験手法 | ELECTRON MICROSCOPY (1.67 Å) | | 主引用文献 | mRNA decoding in human is kinetically and structurally distinct from bacteria.
Nature, 617, 2023
|
|
8G5Z
 
 | | mRNA decoding in human is kinetically and structurally distinct from bacteria (GA state) | | 分子名称: | 1,4-DIAMINOBUTANE, 18S rRNA, 28S rRNA, ... | | 著者 | Holm, M, Natchiar, K.S, Rundlet, E.J, Myasnikov, A.G, Altman, R.B, Blanchard, S.C. | | 登録日 | 2023-02-14 | | 公開日 | 2023-04-19 | | 最終更新日 | 2023-11-15 | | 実験手法 | ELECTRON MICROSCOPY (2.64 Å) | | 主引用文献 | mRNA decoding in human is kinetically and structurally distinct from bacteria.
Nature, 617, 2023
|
|
8G6J
 
 | | mRNA decoding in human is kinetically and structurally distinct from bacteria (GA state 2) | | 分子名称: | (3R,6R,9S,12S,15S,18S,20R,24aR)-6-[(2S)-butan-2-yl]-3,12-bis[(1R)-1-hydroxy-2-methylpropyl]-8,9,11,17,18-pentamethyl-15-[(2S)-2-methylbutyl]hexadecahydropyrido[1,2-a][1,4,7,10,13,16,19]heptaazacyclohenicosine-1,4,7,10,13,16,19(21H)-heptone, (3beta)-O~3~-[(2R)-2,6-dihydroxy-2-(2-methoxy-2-oxoethyl)-6-methylheptanoyl]cephalotaxine, 1,4-DIAMINOBUTANE, ... | | 著者 | Holm, M, Natchiar, K.S, Rundlet, E.J, Myasnikov, A.G, Altman, R.B, Blanchard, S.C. | | 登録日 | 2023-02-15 | | 公開日 | 2023-04-19 | | 最終更新日 | 2023-11-15 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | mRNA decoding in human is kinetically and structurally distinct from bacteria.
Nature, 617, 2023
|
|
8G5Y
 
 | | mRNA decoding in human is kinetically and structurally distinct from bacteria (IC state) | | 分子名称: | 1,4-DIAMINOBUTANE, 18S rRNA, 28S rRNA, ... | | 著者 | Holm, M, Natchiar, K.S, Rundlet, E.J, Myasnikov, A.G, Altman, R.B, Blanchard, S.C. | | 登録日 | 2023-02-14 | | 公開日 | 2023-04-19 | | 最終更新日 | 2023-11-15 | | 実験手法 | ELECTRON MICROSCOPY (2.29 Å) | | 主引用文献 | mRNA decoding in human is kinetically and structurally distinct from bacteria.
Nature, 617, 2023
|
|
8G60
 
 | | mRNA decoding in human is kinetically and structurally distinct from bacteria (CR state) | | 分子名称: | 1,4-DIAMINOBUTANE, 18S rRNA, 28S rRNA, ... | | 著者 | Holm, M, Natchiar, K.S, Rundlet, E.J, Myasnikov, A.G, Altman, R.B, Blanchard, S.C. | | 登録日 | 2023-02-14 | | 公開日 | 2023-04-19 | | 最終更新日 | 2023-11-15 | | 実験手法 | ELECTRON MICROSCOPY (2.54 Å) | | 主引用文献 | mRNA decoding in human is kinetically and structurally distinct from bacteria.
Nature, 617, 2023
|
|
8G61
 
 | | mRNA decoding in human is kinetically and structurally distinct from bacteria (AC state) | | 分子名称: | 1,4-DIAMINOBUTANE, 18S rRNA, 28S rRNA, ... | | 著者 | Holm, M, Natchiar, K.S, Rundlet, E.J, Myasnikov, A.G, Altman, R.B, Blanchard, S.C. | | 登録日 | 2023-02-14 | | 公開日 | 2023-04-19 | | 最終更新日 | 2023-11-15 | | 実験手法 | ELECTRON MICROSCOPY (2.94 Å) | | 主引用文献 | mRNA decoding in human is kinetically and structurally distinct from bacteria.
Nature, 617, 2023
|
|
7YDQ
 
 | | Structure of PfNT1(Y190A)-GFP in complex with GSK4 | | 分子名称: | 5-methyl-N-[2-(2-oxidanylideneazepan-1-yl)ethyl]-2-phenyl-1,3-oxazole-4-carboxamide, Nucleoside transporter 1,Green fluorescent protein | | 著者 | Wang, C, Yu, L.Y, Li, J.L, Ren, R.B, Deng, D. | | 登録日 | 2022-07-04 | | 公開日 | 2023-04-26 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (4.04 Å) | | 主引用文献 | Structural basis of the substrate recognition and inhibition mechanism of Plasmodium falciparum nucleoside transporter PfENT1.
Nat Commun, 14, 2023
|
|
8FI8
 
 | |
8FHV
 
 | | Structure of Lettuce aptamer bound to DFHBI-1T with thallium I ions | | 分子名称: | (5Z)-5-(3,5-difluoro-4-hydroxybenzylidene)-2-methyl-3-(2,2,2-trifluoroethyl)-3,5-dihydro-4H-imidazol-4-one, Lettuce DNA aptamer, MAGNESIUM ION, ... | | 著者 | Passalacqua, L.F.M, Ferre-D'Amare, A.R. | | 登録日 | 2022-12-15 | | 公開日 | 2023-05-10 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Intricate 3D architecture of a DNA mimic of GFP.
Nature, 618, 2023
|
|
8FHZ
 
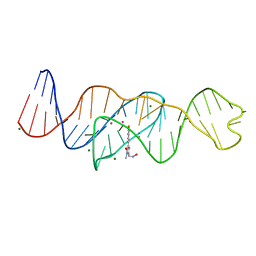 | | Structure of Lettuce aptamer bound to DFHO | | 分子名称: | (5Z)-5-[(3,5-difluoro-4-hydroxyphenyl)methylidene]-2-[(E)-(hydroxyimino)methyl]-3-methyl-3,5-dihydro-4H-imidazol-4-one, Lettuce DNA aptamer, MAGNESIUM ION, ... | | 著者 | Passalacqua, L.F.M, Ferre-D'Amare, A.R. | | 登録日 | 2022-12-15 | | 公開日 | 2023-05-10 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Intricate 3D architecture of a DNA mimic of GFP.
Nature, 618, 2023
|
|
8FI2
 
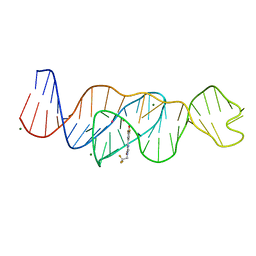 | | Structure of Lettuce C20T bound to DFHBI-1T | | 分子名称: | (5Z)-5-(3,5-difluoro-4-hydroxybenzylidene)-2-methyl-3-(2,2,2-trifluoroethyl)-3,5-dihydro-4H-imidazol-4-one, Lettuce DNA aptamer, MAGNESIUM ION, ... | | 著者 | Passalacqua, L.F.M, Ferre-D'Amare, A.R. | | 登録日 | 2022-12-15 | | 公開日 | 2023-05-10 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Intricate 3D architecture of a DNA mimic of GFP.
Nature, 618, 2023
|
|
8FI1
 
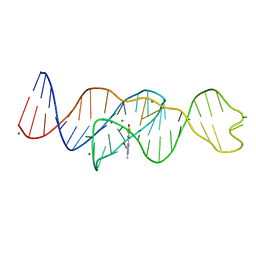 | | Structure of Lettuce C20G bound to DFHO | | 分子名称: | (5Z)-5-[(3,5-difluoro-4-hydroxyphenyl)methylidene]-2-[(E)-(hydroxyimino)methyl]-3-methyl-3,5-dihydro-4H-imidazol-4-one, Lettuce DNA aptamer, MAGNESIUM ION, ... | | 著者 | Passalacqua, L.F.M, Ferre-D'Amare, A.R. | | 登録日 | 2022-12-15 | | 公開日 | 2023-05-10 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Intricate 3D architecture of a DNA mimic of GFP.
Nature, 618, 2023
|
|
8FI0
 
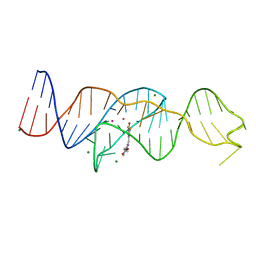 | |
8FI7
 
 | | Structure of Lettuce C20T bound to DFHO | | 分子名称: | (5Z)-5-[(3,5-difluoro-4-hydroxyphenyl)methylidene]-2-[(E)-(hydroxyimino)methyl]-3-methyl-3,5-dihydro-4H-imidazol-4-one, Lettuce DNA aptamer, MAGNESIUM ION, ... | | 著者 | Passalacqua, L.F.M, Ferre-D'Amare, A.R. | | 登録日 | 2022-12-15 | | 公開日 | 2023-05-10 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Intricate 3D architecture of a DNA mimic of GFP.
Nature, 618, 2023
|
|
