7E40
 
 | | Mechanism of Phosphate Sensing and Signaling Revealed by Rice SPX1-PHR2 Complex Structure | | 分子名称: | INOSITOL HEXAKISPHOSPHATE, Protein PHOSPHATE STARVATION RESPONSE 2, SPX domain-containing protein 1,Endolysin | | 著者 | Zhou, J, Hu, Q, Yao, D, Xing, W. | | 登録日 | 2021-02-09 | | 公開日 | 2021-11-10 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Mechanism of phosphate sensing and signaling revealed by rice SPX1-PHR2 complex structure.
Nat Commun, 12, 2021
|
|
1B6W
 
 | |
5JEL
 
 | | Phosphorylated TRIF in complex with IRF-3 | | 分子名称: | Interferon regulatory factor 3, Phosphorylated TRIF peptide | | 著者 | Zhao, B, Li, P. | | 登録日 | 2016-04-18 | | 公開日 | 2016-06-15 | | 最終更新日 | 2016-06-29 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural basis for concerted recruitment and activation of IRF-3 by innate immune adaptor proteins.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5JEO
 
 | | Phosphorylated Rotavirus NSP1 in complex with IRF-3 | | 分子名称: | Interferon regulatory factor 3, PHOSPHATE ION, Rotavirus NSP1 peptide | | 著者 | Zhao, B, Li, P. | | 登録日 | 2016-04-18 | | 公開日 | 2016-06-15 | | 最終更新日 | 2016-06-29 | | 実験手法 | X-RAY DIFFRACTION (1.719 Å) | | 主引用文献 | Structural basis for concerted recruitment and activation of IRF-3 by innate immune adaptor proteins.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6BDA
 
 | | Wild-type I-OnuI bound to A3G substrate (post-cleavage complex) | | 分子名称: | Cleaved Cognate DNA strand, +11 sense, Cleaved cognate DNA strand, ... | | 著者 | Brown, C, Zhang, K, Laforet, M, McMurrough, T, Gloor, G.B, Edgell, D.R, Junop, M. | | 登録日 | 2017-10-22 | | 公開日 | 2018-10-24 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | Wild-type I-OnuI bound to A3G substrate (post-cleavage complex)
To Be Published
|
|
4WWX
 
 | | Crystal structure of the core RAG1/2 recombinase | | 分子名称: | V(D)J recombination-activating protein 1, V(D)J recombination-activating protein 2, ZINC ION | | 著者 | Kim, M.S, Lapkouski, M, Yang, W, Gellert, M. | | 登録日 | 2014-11-12 | | 公開日 | 2015-02-25 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (3.2001 Å) | | 主引用文献 | Crystal structure of the V(D)J recombinase RAG1-RAG2.
Nature, 518, 2015
|
|
4ML0
 
 | | Crystal structure of E.coli DinJ-YafQ complex | | 分子名称: | Predicted antitoxin of YafQ-DinJ toxin-antitoxin system, Predicted toxin of the YafQ-DinJ toxin-antitoxin system, SULFATE ION | | 著者 | Liang, Y.J, Gao, Z.Q, Liu, Q.S, Dong, Y.H. | | 登録日 | 2013-09-06 | | 公開日 | 2014-06-25 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural and Functional Characterization of Escherichia coli Toxin-Antitoxin Complex DinJ-YafQ
J.Biol.Chem., 289, 2014
|
|
1L0B
 
 | | Crystal Structure of rat Brca1 tandem-BRCT region | | 分子名称: | BRCA1 | | 著者 | Joo, W.S, Jeffrey, P.D, Cantor, S.B, Finnin, M.S, Livingston, D.M, Pavletich, N.P. | | 登録日 | 2002-02-08 | | 公開日 | 2002-03-20 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure of the 53BP1 BRCT region bound to p53 and its comparison to the Brca1 BRCT structure.
Genes Dev., 16, 2002
|
|
3MEF
 
 | |
4JZY
 
 | | Crystal structures of Drosophila Cryptochrome | | 分子名称: | AMMONIUM ION, Cryptochrome-1, FLAVIN-ADENINE DINUCLEOTIDE, ... | | 著者 | Czarna, A, Wolf, E. | | 登録日 | 2013-04-03 | | 公開日 | 2013-06-26 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.34 Å) | | 主引用文献 | Structures of Drosophila cryptochrome and mouse cryptochrome1 provide insight into circadian function.
Cell(Cambridge,Mass.), 153, 2013
|
|
4K03
 
 | | Crystal structure of Drosophila Cryprochrome | | 分子名称: | Cryptochrome-1, FLAVIN-ADENINE DINUCLEOTIDE | | 著者 | Berndt, A, Wolf, E. | | 登録日 | 2013-04-03 | | 公開日 | 2013-06-26 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structures of Drosophila cryptochrome and mouse cryptochrome1 provide insight into circadian function.
Cell(Cambridge,Mass.), 153, 2013
|
|
7FSE
 
 | | Crystal Structure of T. maritima reverse gyrase with a minimal latch | | 分子名称: | CHLORIDE ION, DODECAETHYLENE GLYCOL, Reverse gyrase, ... | | 著者 | Rasche, R, Kummel, D, Rudolph, M.G, Klostermeier, D. | | 登録日 | 2023-01-04 | | 公開日 | 2023-05-10 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.89 Å) | | 主引用文献 | Structure of reverse gyrase with a minimal latch that supports ATP-dependent positive supercoiling without specific interactions with the topoisomerase domain.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7FSF
 
 | | CRYSTAL STRUCTURE OF T. MARITIMA REVERSE GYRASE ACTIVE SITE VARIANT Y851F | | 分子名称: | Reverse gyrase, ZINC ION | | 著者 | Rasche, R, Kummel, D, Rudolph, M.G, Klostermeier, D. | | 登録日 | 2023-01-04 | | 公開日 | 2023-05-10 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.77 Å) | | 主引用文献 | Structure of reverse gyrase with a minimal latch that supports ATP-dependent positive supercoiling without specific interactions with the topoisomerase domain.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
5JEK
 
 | | Phosphorylated MAVS in complex with IRF-3 | | 分子名称: | Interferon regulatory factor 3, MAVS peptide | | 著者 | Zhao, B, Li, P. | | 登録日 | 2016-04-18 | | 公開日 | 2016-06-15 | | 最終更新日 | 2016-06-29 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural basis for concerted recruitment and activation of IRF-3 by innate immune adaptor proteins.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4LZ4
 
 | | X-ray structure of the complex between human thrombin and the TBA deletion mutant lacking thymine 3 nucleobase | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, POTASSIUM ION, ... | | 著者 | Pica, A, Russo Krauss, I, Merlino, A, Sica, F. | | 登録日 | 2013-07-31 | | 公開日 | 2014-01-08 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.56 Å) | | 主引用文献 | Dissecting the contribution of thrombin exosite I in the recognition of thrombin binding aptamer.
Febs J., 280, 2013
|
|
6NU2
 
 | | Structural insights into unique features of the human mitochondrial ribosome recycling | | 分子名称: | 12S rRNA, 16S rRNA, 28S ribosomal protein S10, ... | | 著者 | Sharma, M.R, Koripella, R.K, Agrawal, R.K. | | 登録日 | 2019-01-30 | | 公開日 | 2019-04-17 | | 最終更新日 | 2019-12-11 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Structural insights into unique features of the human mitochondrial ribosome recycling.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6UCO
 
 | |
6UCP
 
 | |
3B0C
 
 | | Crystal structure of the chicken CENP-T histone fold/CENP-W complex, crystal form I | | 分子名称: | CITRIC ACID, Centromere protein T, Centromere protein W | | 著者 | Nishino, T, Takeuchi, K, Gascoigne, K.E, Suzuki, A, Hori, T, Oyama, T, Morikawa, K, Cheeseman, I.M, Fukagawa, T. | | 登録日 | 2011-06-08 | | 公開日 | 2012-03-07 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.201 Å) | | 主引用文献 | CENP-T-W-S-X Forms a Unique Centromeric Chromatin Structure with a Histone-like Fold.
Cell(Cambridge,Mass.), 148, 2012
|
|
5JEM
 
 | | Complex of IRF-3 with CBP | | 分子名称: | CREB-binding protein, Interferon regulatory factor 3 | | 著者 | Zhao, B, Li, P. | | 登録日 | 2016-04-18 | | 公開日 | 2016-06-15 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural basis for concerted recruitment and activation of IRF-3 by innate immune adaptor proteins.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1NUO
 
 | | Two RTH Mutants with Impaired Hormone Binding | | 分子名称: | Thyroid hormone receptor beta-1, [4-(4-HYDROXY-3-IODO-PHENOXY)-3,5-DIIODO-PHENYL]-ACETIC ACID | | 著者 | Huber, B.R, Sandler, B, West, B.L, Cunha-Lima, S.T, Nguyen, H.T, Apriletti, J.W, Baxter, J.D, Fletterick, R.J. | | 登録日 | 2003-01-31 | | 公開日 | 2003-04-15 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Two resistance to thyroid hormone mutants with impaired hormone binding
Mol.Endocrinol., 17, 2003
|
|
7CTE
 
 | | Human Origin Recognition Complex, ORC2-5 | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Origin recognition complex subunit 2, Origin recognition complex subunit 3, ... | | 著者 | Cheng, J, Li, N, Wang, X, Hu, J, Zhai, Y, Gao, N. | | 登録日 | 2020-08-18 | | 公開日 | 2021-01-06 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structural insight into the assembly and conformational activation of human origin recognition complex.
Cell Discov, 6, 2020
|
|
6WLG
 
 | |
1ULY
 
 | | Crystal structure analysis of the ArsR homologue DNA-binding protein from P. horikoshii OT3 | | 分子名称: | hypothetical protein PH1932 | | 著者 | Itou, H, Yao, M, Watanabe, N, Tanaka, I. | | 登録日 | 2003-09-17 | | 公開日 | 2004-10-19 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure of the PH1932 protein, a unique archaeal ArsR type winged-HTH transcription factor from Pyrococcus horikoshii OT3
Proteins, 70, 2008
|
|
1B67
 
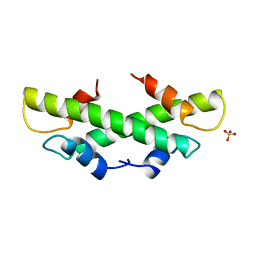 | | CRYSTAL STRUCTURE OF THE HISTONE HMFA FROM METHANOTHERMUS FERVIDUS | | 分子名称: | PROTEIN (HISTONE HMFA), SULFATE ION | | 著者 | Decanniere, K, Sandman, K, Reeve, J.N, Heinemann, U. | | 登録日 | 1999-01-19 | | 公開日 | 2000-01-17 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.48 Å) | | 主引用文献 | Crystal structures of recombinant histones HMfA and HMfB from the hyperthermophilic archaeon Methanothermus fervidus.
J.Mol.Biol., 303, 2000
|
|
