8UW3
 
 | | Human LINE-1 retrotransposon ORF2 protein engaged with template RNA in elongation state | | 分子名称: | Complementary DNA, LINE-1 retrotransposable element ORF2 protein, THYMIDINE-5'-TRIPHOSPHATE, ... | | 著者 | Thawani, A, Florez Ariza, A.J, Collins, K, Nogales, E. | | 登録日 | 2023-11-06 | | 公開日 | 2023-12-20 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Template and target-site recognition by human LINE-1 in retrotransposition.
Nature, 626, 2024
|
|
8PYX
 
 | |
8PPP
 
 | |
8PYY
 
 | |
7S7I
 
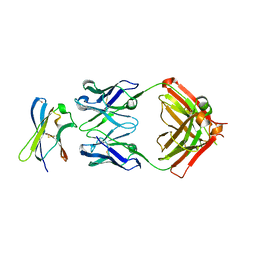 | |
2MQ8
 
 | | Solution NMR Structure of De novo designed protein LFR1 1 with ferredoxin fold, Northeast Structural Genomics Consortium (NESG) Target OR414 | | 分子名称: | De novo designed protein LFR1 | | 著者 | Liu, G, Lin, Y, Koga, N, Koga, R, Xiao, R, Janjua, H, Pederson, K, Acton, T.B, Kornhaber, G, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2014-06-12 | | 公開日 | 2014-08-20 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Control over overall shape and size in de novo designed proteins.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
7QUL
 
 | | Alcohol Dehydrogenase from Thauera aromatica K319A/K320A mutant | | 分子名称: | 1,2-ETHANEDIOL, 6-hydroxycyclohex-1-ene-1-carbonyl-CoA dehydrogenase, ZINC ION | | 著者 | Petchey, M.L, Stark, F, Ansorge-Schumacher, M, Grogan, G. | | 登録日 | 2022-01-18 | | 公開日 | 2022-08-17 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Advanced Insights into Catalytic and Structural Features of the Zinc-Dependent Alcohol Dehydrogenase from Thauera aromatica.
Chembiochem, 23, 2022
|
|
7QUY
 
 | | Alcohol Dehydrogenase from Thauera aromatica complexed with NADH | | 分子名称: | 6-hydroxycyclohex-1-ene-1-carbonyl-CoA dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, ... | | 著者 | Petchey, M.L, Stark, F, Ansorge-Schumacher, M, Grogan, G. | | 登録日 | 2022-01-19 | | 公開日 | 2022-08-17 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Advanced Insights into Catalytic and Structural Features of the Zinc-Dependent Alcohol Dehydrogenase from Thauera aromatica.
Chembiochem, 23, 2022
|
|
7QZN
 
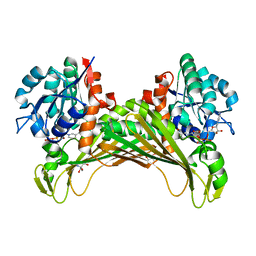 | | Amine Dehydrogenase from Cystobacter fuscus (CfusAmDH) W145A mutant with NAD+ | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Amine Dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Bennett, M, Ducrot, L, Vaxelaire-Vergne, C, Grogan, G. | | 登録日 | 2022-01-31 | | 公開日 | 2022-12-07 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.64 Å) | | 主引用文献 | Cover Feature: Expanding the Substrate Scope of Native Amine Dehydrogenases through In Silico Structural Exploration and Targeted Protein Engineering (ChemCatChem 22/2022)
Chemcatchem, 14, 2022
|
|
7QZL
 
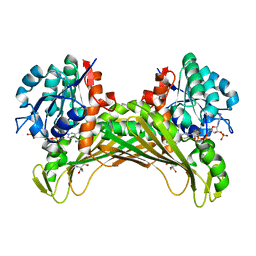 | | Amine Dehydrogenase from Cystobacter fuscus (CfusAmDH) W145A mutant with NADP+ and pentylamine | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, AMYLAMINE, Amine Dehydrogenase, ... | | 著者 | Bennett, M, Ducrot, L, Vaxelaire-Vergne, C, Grogan, G. | | 登録日 | 2022-01-31 | | 公開日 | 2022-12-07 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Cover Feature: Expanding the Substrate Scope of Native Amine Dehydrogenases through In Silico Structural Exploration and Targeted Protein Engineering (ChemCatChem 22/2022)
Chemcatchem, 14, 2022
|
|
7R09
 
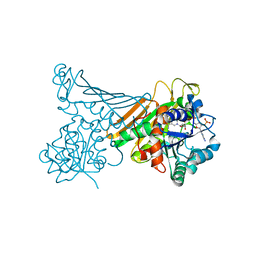 | | Amine Dehydrogenase MATOUAmDH2 in complex with NADP+ and Cyclohexylamine | | 分子名称: | Amine Dehydrogenase, CYCLOHEXYLAMMONIUM ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Bennett, M, Ducrot, L, Vaxelaire-Vergne, C, Grogan, G. | | 登録日 | 2022-02-01 | | 公開日 | 2022-04-06 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.08 Å) | | 主引用文献 | Structure and Mutation of the Native Amine Dehydrogenase MATOUAmDH2.
Chembiochem, 23, 2022
|
|
7KBQ
 
 | |
3MCH
 
 | | Crystal structure of the molybdopterin biosynthesis enzyme MoaB (TTHA0341) from thermus theromophilus HB8 | | 分子名称: | 1,2-ETHANEDIOL, Molybdopterin biosynthesis enzyme, MoaB | | 著者 | Jeyakanthan, J, Kanaujia, S.P, Sekar, K, Baba, S, Ebihara, A, Kuramitsu, S, Shinkai, A, Shiro, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2010-03-29 | | 公開日 | 2011-01-19 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.64 Å) | | 主引用文献 | Crystal structures, dynamics and functional implications of molybdenum-cofactor biosynthesis protein MogA from two thermophilic organisms
Acta Crystallogr.,Sect.F, 67, 2011
|
|
1V5X
 
 | |
8FZ8
 
 | | Structure of cytochrome P450sky2 | | 分子名称: | Cytochrome P450, OCTANOIC ACID (CAPRYLIC ACID), PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Murarka, V.C, Poulos, T.L. | | 登録日 | 2023-01-27 | | 公開日 | 2024-02-07 | | 最終更新日 | 2024-04-10 | | 実験手法 | X-RAY DIFFRACTION (1.43 Å) | | 主引用文献 | Biosynthesis of a new skyllamycin in Streptomyces nodosus : a cytochrome P450 forms an epoxide in the cinnamoyl chain.
Org.Biomol.Chem., 22, 2024
|
|
4USR
 
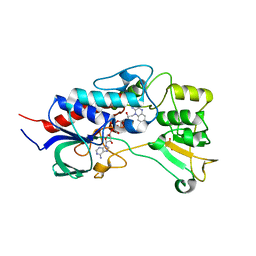 | | Structure of flavin-containing monooxygenase from Pseudomonas stutzeri NF13 | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, MONOOXYGENASE | | 著者 | Jensen, C.N, Ali, S.T, Allen, M.J, Grogan, G. | | 登録日 | 2014-07-11 | | 公開日 | 2014-10-01 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.83 Å) | | 主引用文献 | Exploring Nicotinamide Cofactor Promiscuity in Nad(P)H-Dependent Flavin Containing Monooxygenases (Fmos) Using Natural Variation within the Phosphate Binding Loop. Structure and Activity of Fmos from Cellvibrio Sp. Br and Pseudomonas Stutzeri NF13
J.Mol.Catal., 109, 2014
|
|
6FL8
 
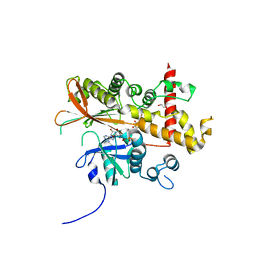 | | Inositol 1,3,4,5,6-pentakisphosphate 2-kinase from A. thaliana in complex with purpurogallin and ADP | | 分子名称: | 1,2-ETHANEDIOL, 2,3,4,6-tetrahydroxy-5H-benzo[7]annulen-5-one, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | 著者 | Whitfield, H.L, Brearley, C.A, Hemmings, A.M. | | 登録日 | 2018-01-25 | | 公開日 | 2018-09-12 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | A Fluorescent Probe Identifies Active Site Ligands of Inositol Pentakisphosphate 2-Kinase.
J. Med. Chem., 61, 2018
|
|
8GUJ
 
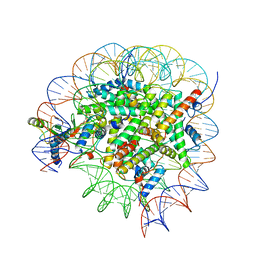 | | Bre1-nucleosome complex (Model II) | | 分子名称: | DNA (147-mer), E3 ubiquitin-protein ligase BRE1A, E3 ubiquitin-protein ligase BRE1B, ... | | 著者 | Onishi, S, Sato, K, Hamada, K, Nishizawa, T, Nureki, O, Ogata, K, Sengoku, T. | | 登録日 | 2022-09-12 | | 公開日 | 2023-09-20 | | 最終更新日 | 2024-04-03 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Structure of the human Bre1 complex bound to the nucleosome.
Nat Commun, 15, 2024
|
|
8GUK
 
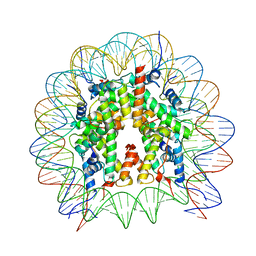 | | Human nucleosome core particle (free form) | | 分子名称: | DNA (147-mer), Histone H2A type 1, Histone H2B type 1-J, ... | | 著者 | Onishi, S, Sato, K, Nishizawa, T, Nureki, O, Ogata, K, Sengoku, T. | | 登録日 | 2022-09-12 | | 公開日 | 2023-09-20 | | 最終更新日 | 2024-04-03 | | 実験手法 | ELECTRON MICROSCOPY (2.51 Å) | | 主引用文献 | Structure of the human Bre1 complex bound to the nucleosome.
Nat Commun, 15, 2024
|
|
8GUI
 
 | | Bre1-nucleosome complex (Model I) | | 分子名称: | DNA (147-mer), E3 ubiquitin-protein ligase BRE1A, E3 ubiquitin-protein ligase BRE1B, ... | | 著者 | Onishi, S, Hamada, K, Sato, K, Nishizawa, T, Nureki, O, Ogata, K, Sengoku, T. | | 登録日 | 2022-09-12 | | 公開日 | 2023-09-20 | | 最終更新日 | 2024-04-03 | | 実験手法 | ELECTRON MICROSCOPY (2.81 Å) | | 主引用文献 | Structure of the human Bre1 complex bound to the nucleosome.
Nat Commun, 15, 2024
|
|
6FP5
 
 | | Crystal structure of ZAD-domain of CG2712 protein from D.melanogaster | | 分子名称: | CG2712, GLYCEROL, ZINC ION | | 著者 | Boyko, K.M, Nikolaeva, A.Y, Bonchuk, A.N, Kachalova, G.S, Georgiev, P.G, Popov, V.O. | | 登録日 | 2018-02-09 | | 公開日 | 2019-08-21 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural basis of diversity and homodimerization specificity of zinc-finger-associated domains in Drosophila.
Nucleic Acids Res., 49, 2021
|
|
6OGP
 
 | | X-ray crystal structure of wild type HIV-1 protease in complex with GRL-063 | | 分子名称: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl {(2S,3R)-1-(3,5-difluorophenyl)-3-hydroxy-4-[(2-methylpropyl)({2-[(propan-2-yl)amino]-1,3-benzoxazol-6-yl}sulfonyl)amino]butan-2-yl}carbamate, 1,2-ETHANEDIOL, Protease | | 著者 | Bulut, H, Hattori, S.I, Aoki-Ogata, H, Hayashi, H, Aoki, M, Ghosh, A.K, Mitsuya, H. | | 登録日 | 2019-04-03 | | 公開日 | 2020-04-08 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.53 Å) | | 主引用文献 | Single atom changes in newly synthesized HIV protease inhibitors reveal structural basis for extreme affinity, high genetic barrier, and adaptation to the HIV protease plasticity.
Sci Rep, 10, 2020
|
|
6OGT
 
 | | X-ray crystal structure of darunavir-resistant HIV-1 protease (P51) in complex with GRL-001 | | 分子名称: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl [(2S,3R)-4-[{[2-(cyclopropylamino)-1,3-benzothiazol-6-yl]sulfonyl}(2-methylpropyl)amino]-1-(3-fluorophenyl)-3-hydroxybutan-2-yl]carbamate, 1,2-ETHANEDIOL, GLYCEROL, ... | | 著者 | Bulut, H, Hattori, S.I, Aoki-Ogata, H, Hayashi, H, Aoki, M, Ghosh, A.K, Mitsuya, H. | | 登録日 | 2019-04-03 | | 公開日 | 2020-04-08 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.21 Å) | | 主引用文献 | Single atom changes in newly synthesized HIV protease inhibitors reveal structural basis for extreme affinity, high genetic barrier, and adaptation to the HIV protease plasticity.
Sci Rep, 10, 2020
|
|
6OGL
 
 | | X-ray crystal structure of darunavir-resistant HIV-1 protease (P51) in complex with GRL-003 | | 分子名称: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl [(2S,3R)-4-[{[2-(cyclopropylamino)-1,3-benzothiazol-6-yl]sulfonyl}(2-methylpropyl)amino]-1-(4-fluorophenyl)-3-hydroxybutan-2-yl]carbamate, 1,2-ETHANEDIOL, GLYCEROL, ... | | 著者 | Bulut, H, Hattori, S.I, Aoki-Ogata, H, Hayashi, H, Aoki, M, Ghosh, A.K, Mitsuya, H. | | 登録日 | 2019-04-02 | | 公開日 | 2020-04-08 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.21 Å) | | 主引用文献 | Single atom changes in newly synthesized HIV protease inhibitors reveal structural basis for extreme affinity, high genetic barrier, and adaptation to the HIV protease plasticity.
Sci Rep, 10, 2020
|
|
6OGV
 
 | | X-ray crystal structure of darunavir-resistant HIV-1 protease (P30) in apo state | | 分子名称: | Protease | | 著者 | Bulut, H, Hattori, S.I, Aoki-Ogata, H, Hayashi, H, Aoki, M, Ghosh, A.K, Mitsuya, H. | | 登録日 | 2019-04-03 | | 公開日 | 2020-04-08 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.57 Å) | | 主引用文献 | Novel HIV-1 protease inhibitors, GRL-142 and its analogs, markedly adapt to the structural plasticity of HIV-1 protease and exerts extreme potency with high genetic barrier
To Be Published
|
|
