7JN1
 
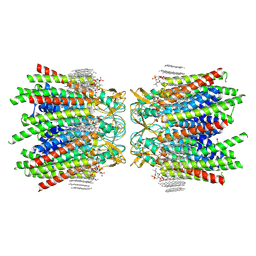 | | Sheep Connexin-46 at 2.5 angstroms resolution, Lipid Class 3 | | 分子名称: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Gap junction alpha-3 protein | | 著者 | Flores, J.A, Haddad, B.G, Dolan, K.D, Myers, J.B, Yoshioka, C.C, Copperman, J, Zuckerman, D.M, Reichow, S.L. | | 登録日 | 2020-08-03 | | 公開日 | 2020-09-09 | | 実験手法 | ELECTRON MICROSCOPY (2.5 Å) | | 主引用文献 | Connexin-46/50 in a dynamic lipid environment resolved by CryoEM at 1.9 angstrom.
Nat Commun, 11, 2020
|
|
2NPZ
 
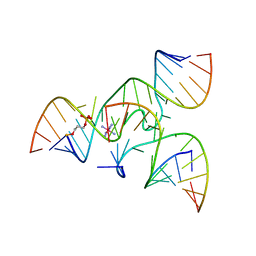 | | Crystal structure of junctioned hairpin ribozyme incorporating synthetic propyl linker | | 分子名称: | 5'-R(*CP*GP*GP*UP*GP*AP*GP*AP*AP*GP*GP*G)-3', 5'-R(*GP*GP*CP*AP*GP*AP*GP*AP*AP*AP*CP*AP*CP*AP*CP*GP*A)-3', 5'-R(*UP*CP*CP*CP*AP*GP*UP*CP*CP*AP*CP*CP*G)-3', ... | | 著者 | MacElrevey, C, Krucinska, J, Wedekind, J.E. | | 登録日 | 2006-10-30 | | 公開日 | 2007-08-14 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (3.35 Å) | | 主引用文献 | A posteriori design of crystal contacts to improve the X-ray diffraction properties of a small RNA enzyme.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
5L4N
 
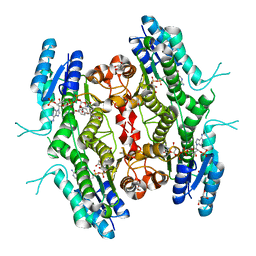 | | Leishmania major Pteridine reductase 1 (PTR1) in complex with compound 1 | | 分子名称: | (2~{R})-2-(3-hydroxyphenyl)-6-oxidanyl-2,3-dihydrochromen-4-one, 1,2-ETHANEDIOL, ACETATE ION, ... | | 著者 | Dello Iacono, L, Di Pisa, F, Pozzi, C, Landi, G, Mangani, S. | | 登録日 | 2016-05-26 | | 公開日 | 2017-03-22 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Chroman-4-One Derivatives Targeting Pteridine Reductase 1 and Showing Anti-Parasitic Activity.
Molecules, 22, 2017
|
|
2LL2
 
 | | Structure of the Cx43 C-terminal domain bound to tubulin | | 分子名称: | Gap junction alpha-1 protein | | 著者 | Saidi Brikci-Nigassa, A, Clement, M, Ha-Duong, T, Benmansour, K, Adjadj, E, Ziani, L, Pastre, D, Curmi, P.A. | | 登録日 | 2011-10-26 | | 公開日 | 2012-06-20 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Phosphorylation controls the interaction of the connexin43 C-terminal domain with tubulin and microtubules.
Biochemistry, 51, 2012
|
|
8IVU
 
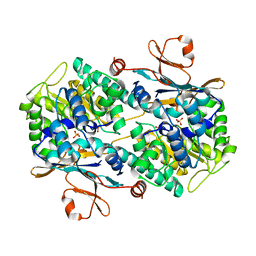 | | Crystal Structure of Human NAMPT in complex with A4276 | | 分子名称: | N-[[4-(6-methyl-1,3-benzoxazol-2-yl)phenyl]methyl]pyridine-3-carboxamide, Nicotinamide phosphoribosyltransferase, PHOSPHATE ION | | 著者 | Kang, B.G, Cha, S.S. | | 登録日 | 2023-03-28 | | 公開日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.09000921 Å) | | 主引用文献 | Discovery of a novel NAMPT inhibitor that selectively targets NAPRT-deficient EMT-subtype cancer cells and alleviates chemotherapy-induced peripheral neuropathy.
Theranostics, 13, 2023
|
|
7KE9
 
 | |
7KEB
 
 | |
7KDH
 
 | |
7KDI
 
 | |
7KEA
 
 | |
7KDG
 
 | |
7KDL
 
 | |
7KE8
 
 | |
7KE4
 
 | |
7KDJ
 
 | |
7KDK
 
 | |
7KEC
 
 | |
7KE6
 
 | |
7KE7
 
 | |
5X1E
 
 | |
4BAE
 
 | | Optimisation of pyrroleamides as mycobacterial GyrB ATPase inhibitors: Structure Activity Relationship and in vivo efficacy in the mouse model of tuberculosis | | 分子名称: | 2-[(3S,4R)-4-[(3-bromanyl-4-chloranyl-5-methyl-1H-pyrrol-2-yl)carbonylamino]-3-methoxy-piperidin-1-yl]-4-(2-methyl-1,2,4-triazol-3-yl)-1,3-thiazole-5-carboxylic acid, CALCIUM ION, DNA GYRASE SUBUNIT B, ... | | 著者 | Read, J.A, Gingell, H.G, Madhavapeddi, P. | | 登録日 | 2012-09-13 | | 公開日 | 2013-10-30 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Optimization of Pyrrolamides as Mycobacterial Gyrb ATPase Inhibitors: Structure Activity Relationship and in Vivo Efficacy in the Mouse Model of Tuberculosis.
Antimicrob.Agents Chemother., 58, 2014
|
|
8IAH
 
 | | Structure of mammalian spectrin-actin junctional complex of membrane skeleton, State I, Global map | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Actin, cytoplasmic 1, ... | | 著者 | Li, N, Chen, S, Gao, N. | | 登録日 | 2023-02-08 | | 公開日 | 2023-05-03 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structural basis of membrane skeleton organization in red blood cells.
Cell, 186, 2023
|
|
2NPY
 
 | | Crystal Structure of a junctioned hairpin ribozyme incorporating 9atom linker and 2'-deoxy 2'-amino U at A-1 | | 分子名称: | 2-[2-(2-HYDROXYETHOXY)ETHOXY]ETHYL DIHYDROGEN PHOSPHATE, 5'-R(*CP*GP*GP*UP*GP*AP*GP*AP*AP*GP*GP*G)-3', 5'-R(*GP*GP*CP*AP*GP*AP*GP*AP*AP*AP*CP*AP*CP*AP*CP*GP*A)-3', ... | | 著者 | MacElrevey, C, Krucinska, J, Wedekind, J.E. | | 登録日 | 2006-10-30 | | 公開日 | 2007-08-14 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | A posteriori design of crystal contacts to improve the X-ray diffraction properties of a small RNA enzyme.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
8IAI
 
 | | Structure of mammalian spectrin-actin junctional complex of membrane skeleton, State II, Global map | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Actin, cytoplasmic 1, ... | | 著者 | Li, N, Chen, S, Gao, N. | | 登録日 | 2023-02-08 | | 公開日 | 2023-05-03 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structural basis of membrane skeleton organization in red blood cells.
Cell, 186, 2023
|
|
2H5X
 
 | | RuvA from Mycobacterium tuberculosis | | 分子名称: | GLYCEROL, Holliday junction ATP-dependent DNA helicase ruvA | | 著者 | Prabu, J.R, Thamotharan, S, Khanduja, J.S, Alipio, E.Z, Kim, C.Y, Waldo, G.S, Terwilliger, T.C, Segelke, B, Lekin, T, Toppani, D, Hung, L.W, Yu, M, Bursey, E, Muniyappa, K, Chandra, N.R, Vijayan, M. | | 登録日 | 2006-05-28 | | 公開日 | 2006-08-15 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structure of Mycobacterium tuberculosis RuvA, a protein involved in recombination.
ACTA CRYSTALLOGR.,SECT.F, 62, 2006
|
|
