4L20
 
 | |
3F0C
 
 | | Crystal structure of transcriptional regulator from Cytophaga hutchinsonii ATCC 33406 | | 分子名称: | SULFATE ION, Transcriptional regulator | | 著者 | Nocek, B, Maltseva, N, Tan, K, Abdullah, J, Eschenfeldt, W, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2008-10-24 | | 公開日 | 2008-11-11 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (2.96 Å) | | 主引用文献 | Crystal structure of transcriptional regulator from Cytophaga hutchinsonii ATCC 33406
To be Published
|
|
7F8Y
 
 | | Crystal structure of the cholecystokinin receptor CCKAR in complex with devazepide | | 分子名称: | N-[(3S)-1-methyl-2-oxidanylidene-5-phenyl-3H-1,4-benzodiazepin-3-yl]-1H-indole-2-carboxamide, fusion protein of Cholecystokinin receptor type A and Endolysin | | 著者 | Zhang, X, He, C, Wang, M, Zhou, Q, Yang, D, Zhu, Y, Wu, B, Zhao, Q. | | 登録日 | 2021-07-02 | | 公開日 | 2021-10-13 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structures of the human cholecystokinin receptors bound to agonists and antagonists.
Nat.Chem.Biol., 17, 2021
|
|
3FH4
 
 | | Crystal Structure of Recombinant Vibrio proteolyticus aminopeptidase | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Bacterial leucyl aminopeptidase, SODIUM ION, ... | | 著者 | Yong, W, Kim, J.-J.P, Hartley, M, Bennett, B. | | 登録日 | 2008-12-08 | | 公開日 | 2009-11-17 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Heterologous expression and purification of Vibrio proteolyticus (Aeromonas proteolytica) aminopeptidase: a rapid protocol
Protein Expr.Purif., 66, 2009
|
|
7F8U
 
 | | Crystal structure of the cholecystokinin receptor CCKAR in complex with lintitript | | 分子名称: | 2-[2-[[4-(2-chlorophenyl)-1,3-thiazol-2-yl]carbamoyl]indol-1-yl]ethanoic acid, Fusion protein of Cholecystokinin receptor type A and Endolysin | | 著者 | Zhang, X, He, C, Wang, M, Zhou, Q, Yang, D, Zhu, Y, Wu, B, Zhao, Q. | | 登録日 | 2021-07-02 | | 公開日 | 2021-10-13 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structures of the human cholecystokinin receptors bound to agonists and antagonists.
Nat.Chem.Biol., 17, 2021
|
|
4U4I
 
 | | Megavirus chilensis superoxide dismutase | | 分子名称: | Cu/Zn superoxide dismutase | | 著者 | Lartigue, A, Claverie, J.-M, Burlat, B, Coutard, B, Abergel, C. | | 登録日 | 2014-07-23 | | 公開日 | 2014-11-05 | | 最終更新日 | 2014-12-24 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | The megavirus chilensis cu,zn-superoxide dismutase: the first viral structure of a typical cellular copper chaperone-independent hyperstable dimeric enzyme.
J.Virol., 89, 2015
|
|
1RPZ
 
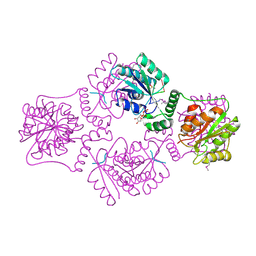 | | T4 POLYNUCLEOTIDE KINASE BOUND TO 5'-TGCAC-3' SSDNA | | 分子名称: | 5'-D(*TP*GP*CP*AP*C)-3', ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, ... | | 著者 | Eastberg, J.H, Pelletier, J, Stoddard, B.L. | | 登録日 | 2003-12-03 | | 公開日 | 2004-02-17 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Recognition of DNA substrates by T4 bacteriophage polynucleotide kinase.
Nucleic Acids Res., 32, 2004
|
|
4ZEK
 
 | |
7FFO
 
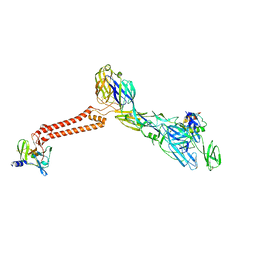 | | Cryo-EM structure of VEEV VLP at the 5-fold axes | | 分子名称: | Capsid protein, Spike glycoprotein E1, Spike glycoprotein E2, ... | | 著者 | Zhang, X, Xiang, Y, Ma, J, Ma, B, Huang, C. | | 登録日 | 2021-07-23 | | 公開日 | 2021-10-20 | | 最終更新日 | 2024-10-16 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structure of Venezuelan equine encephalitis virus with its receptor LDLRAD3.
Nature, 598, 2021
|
|
4LNY
 
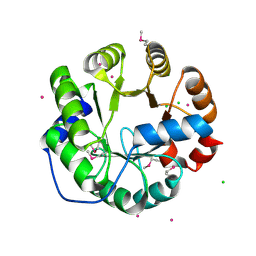 | | Crystal Structure of Engineered Protein, Northeast Structural Genomics Consortium Target OR422 | | 分子名称: | CADMIUM ION, CHLORIDE ION, Engineered Protein OR422 | | 著者 | Vorobiev, S, Su, M, Bjelic, S, Kipnis, Y, Wang, L, Sahdev, S, Xiao, R, Maglaqui, M, Kogan, S, Baker, D, Everett, J.K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2013-07-12 | | 公開日 | 2013-08-07 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.929 Å) | | 主引用文献 | Crystal Structure of Engineered Protein OR422.
To be Published
|
|
2NNE
 
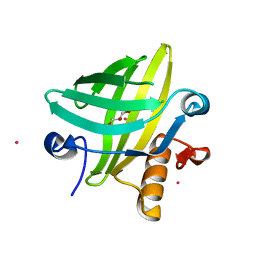 | | The Structural Identification of the Interaction Site and Functional State of RBP for its Membrane Receptor | | 分子名称: | CADMIUM ION, GLYCEROL, Major urinary protein 2 | | 著者 | Redondo, C, Bingham, R.J, Vouropoulou, M, Homans, S.W, Findlay, J.B. | | 登録日 | 2006-10-24 | | 公開日 | 2007-10-30 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Identification of the retinol-binding protein (RBP) interaction site and functional state of RBPs for the membrane receptor.
Faseb J., 22, 2008
|
|
2NQ7
 
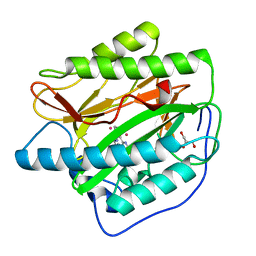 | | Crystal structure of type 1 human methionine aminopeptidase in complex with 3-(2,2-Dimethylpropionylamino)pyridine-2-carboxylic acid thiazole-2-ylamide | | 分子名称: | 3-[(2,2-DIMETHYLPROPANOYL)AMINO]-N-1,3-THIAZOL-2-YLPYRIDINE-2-CARBOXAMIDE, COBALT (II) ION, GLYCEROL, ... | | 著者 | Addlagatta, A, Matthews, B.W. | | 登録日 | 2006-10-30 | | 公開日 | 2006-11-21 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Elucidation of the function of type 1 human methionine aminopeptidase during cell cycle progression.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
5HCR
 
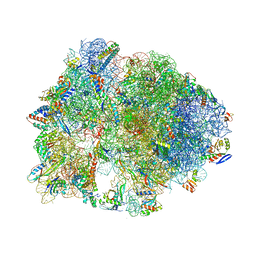 | | Crystal structure of antimicrobial peptide Oncocin 10wt bound to the Thermus thermophilus 70S ribosome | | 分子名称: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Gagnon, M.G, Roy, R.N, Lomakin, I.B, Florin, T, Mankin, A.S, Steitz, T.A. | | 登録日 | 2016-01-04 | | 公開日 | 2016-04-06 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structures of proline-rich peptides bound to the ribosome reveal a common mechanism of protein synthesis inhibition.
Nucleic Acids Res., 44, 2016
|
|
7FFN
 
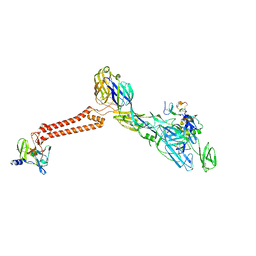 | | Cryo-EM structure of VEEV VLP-LDLRAD3-D1 complex at the 5-fold axes | | 分子名称: | CALCIUM ION, Capsid protein, Low-density lipoprotein receptor class A domain-containing protein 3, ... | | 著者 | Zhang, X, Xiang, Y, Ma, J, Ma, B, Huang, C. | | 登録日 | 2021-07-23 | | 公開日 | 2021-10-20 | | 最終更新日 | 2021-11-17 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structure of Venezuelan equine encephalitis virus with its receptor LDLRAD3.
Nature, 598, 2021
|
|
6RXZ
 
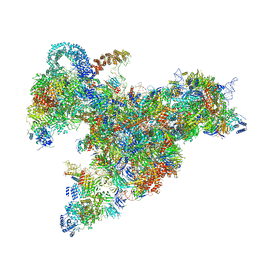 | | Cryo-EM structure of the 90S pre-ribosome (Kre33-Noc4) from Chaetomium thermophilum, state b | | 分子名称: | 35S ribosomal RNA, 40S ribosomal protein S11-like protein, 40S ribosomal protein S13-like protein, ... | | 著者 | Cheng, J, Kellner, N, Griesel, S, Berninghausen, O, Beckmann, R, Hurt, E. | | 登録日 | 2019-06-10 | | 公開日 | 2019-08-14 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | Thermophile 90S Pre-ribosome Structures Reveal the Reverse Order of Co-transcriptional 18S rRNA Subdomain Integration.
Mol.Cell, 75, 2019
|
|
3CS1
 
 | | Flagellar Calcium-binding Protein (FCaBP) from T. cruzi | | 分子名称: | Flagellar calcium-binding protein | | 著者 | Ames, J.B, Ladner, J.E, Wingard, J.N, Robinson, H, Fisher, A. | | 登録日 | 2008-04-08 | | 公開日 | 2008-06-24 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural Insights into Membrane Targeting by the Flagellar Calcium-binding Protein (FCaBP), a Myristoylated and Palmitoylated Calcium Sensor in Trypanosoma cruzi.
J.Biol.Chem., 283, 2008
|
|
3CMZ
 
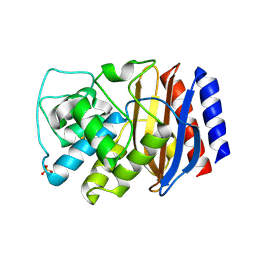 | | TEM-1 Class-A beta-lactamase L201P mutant apo structure | | 分子名称: | Beta-lactamase TEM, PHOSPHATE ION | | 著者 | Marciano, D.C, Wang, X, Wang, J, Chen, Y, Thomas, V.L, Shoichet, B.K, Palzkill, T. | | 登録日 | 2008-03-24 | | 公開日 | 2008-11-25 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | Genetic and structural characterization of an L201P global suppressor substitution in TEM-1 beta-lactamase
J.Mol.Biol., 384, 2008
|
|
7FHZ
 
 | |
5O4W
 
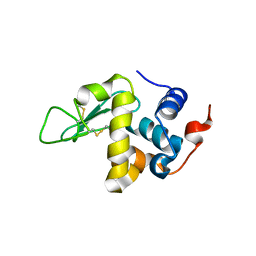 | | Protein structure determination by electron diffraction using a single three-dimensional nanocrystal | | 分子名称: | Lysozyme C | | 著者 | Clabbers, M.T.B, van Genderen, E, Wan, W, Wiegers, E.L, Gruene, T, Abrahams, J.P. | | 登録日 | 2017-05-31 | | 公開日 | 2017-08-23 | | 最終更新日 | 2024-01-17 | | 実験手法 | ELECTRON CRYSTALLOGRAPHY (2.11 Å) | | 主引用文献 | Protein structure determination by electron diffraction using a single three-dimensional nanocrystal.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
4U6S
 
 | | CtBP1 in complex with substrate phenylpyruvate | | 分子名称: | 3-PHENYLPYRUVIC ACID, C-terminal-binding protein 1, CALCIUM ION, ... | | 著者 | Hilbert, B.J, Morris, B.L, Ellis, K.C, Paulsen, J.L, Schiffer, C.A, Grossman, S.R, Royer Jr, W.E. | | 登録日 | 2014-07-29 | | 公開日 | 2015-02-11 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure-Guided Design of a High Affinity Inhibitor to Human CtBP.
Acs Chem.Biol., 10, 2015
|
|
7FI2
 
 | |
1RHZ
 
 | | The structure of a protein conducting channel | | 分子名称: | Preprotein translocase secE subunit, Preprotein translocase secY subunit, SecBeta | | 著者 | van den Berg, B, Clemons Jr, W.M, Collinson, I, Modis, Y, Hartmann, E, Harrison, S.C, Rapoport, T.A. | | 登録日 | 2003-11-15 | | 公開日 | 2004-01-06 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | X-ray structure of a protein-conducting channel.
Nature, 427, 2004
|
|
2NOC
 
 | | Solution Structure of Putative periplasmic protein: Northest Structural Genomics Target StR106 | | 分子名称: | Putative periplasmic protein | | 著者 | Zhang, Q, Liu, G, Wang, H, Nwosu, C, Cunningham, K, Ma, L.C, Xiao, R, Liu, J, Baran, M.C, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2006-10-25 | | 公開日 | 2006-11-28 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution Structure of Putative periplasmic protein: Northest Structural Genomics Target StR106
To be Published
|
|
1R9Z
 
 | | Bacterial cytosine deaminase D314S mutant. | | 分子名称: | Cytosine deaminase, FE (III) ION, GLYCEROL, ... | | 著者 | Mahan, S.D, Ireton, G.C, Stoddard, B.L, Black, M.E. | | 登録日 | 2003-10-31 | | 公開日 | 2004-10-05 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.32 Å) | | 主引用文献 | Random mutagenesis and selection of Escherichia coli cytosine deaminase for cancer gene therapy.
Protein Eng.Des.Sel., 17, 2004
|
|
6S40
 
 | | Fragment AZ-001 binding at the p53pT387/14-3-3 sigma interface and additional sites | | 分子名称: | 14-3-3 protein sigma, 4-chloranyl-1-benzothiophene-2-carboximidamide, CALCIUM ION, ... | | 著者 | Leysen, S, Guillory, X, Wolter, M, Genet, S, Somsen, B, Patel, J, Castaldi, P, Ottmann, C. | | 登録日 | 2019-06-26 | | 公開日 | 2020-06-17 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Fragment-based Differential Targeting of PPI Stabilizer Interfaces.
J.Med.Chem., 63, 2020
|
|
