6S0T
 
 | | The crystal structure of kanamycin B dioxygenase (KanJ) from Streptomyces kanamyceticus in complex with nickel, sulfate, soaked with iodide | | 分子名称: | IODIDE ION, Kanamycin B dioxygenase, NICKEL (II) ION, ... | | 著者 | Mrugala, B, Porebski, P.J, Niedzialkowska, E, Cymborowski, M.T, Minor, W, Borowski, T. | | 登録日 | 2019-06-18 | | 公開日 | 2020-07-08 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | A study on the structure, mechanism, and biochemistry of kanamycin B dioxygenase (KanJ)-an enzyme with a broad range of substrates.
Febs J., 288, 2021
|
|
6SGF
 
 | | Molecular insight into a new low affinity xylan binding module CBM86, from the xylanolytic gut symbiont Roseburia intestinalis. | | 分子名称: | Beta-xylanase, CADMIUM ION, CHLORIDE ION, ... | | 著者 | Ejby, M, Abou Hachem, M, Leth, M.L, Guskov, A, Slotboom, D. | | 登録日 | 2019-08-04 | | 公開日 | 2019-11-20 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (1.756 Å) | | 主引用文献 | Molecular insight into a new low-affinity xylan binding module from the xylanolytic gut symbiont Roseburia intestinalis.
Febs J., 287, 2020
|
|
6THF
 
 | | Crystal structure of two-domain Cu nitrite reductase from Bradyrhizobium sp. ORS 375 | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, COPPER (II) ION, Copper-containing nitrite reductase, ... | | 著者 | Sasaki, D, Watanabe, T.F, Eady, R.R, Garratt, R.C, Antonyuk, S.V, Hasnain, S.S. | | 登録日 | 2019-11-20 | | 公開日 | 2020-04-22 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.47 Å) | | 主引用文献 | Reverse protein engineering of a novel 4-domain copper nitrite reductase reveals functional regulation by protein-protein interaction.
Febs J., 288, 2021
|
|
6R82
 
 | |
6S1U
 
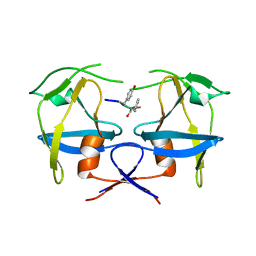 | | Crystal structure of dimeric M-PMV protease C7A/D26N/C106A mutant in complex with inhibitor | | 分子名称: | Gag-Pro-Pol polyprotein, PRO-0A1-VAL-PSA-ALA-MET-THR | | 著者 | Wosicki, S, Gilski, M, Jaskolski, M, Zabranska, H, Pichova, I. | | 登録日 | 2019-06-19 | | 公開日 | 2019-10-16 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Comparison of a retroviral protease in monomeric and dimeric states.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6S1W
 
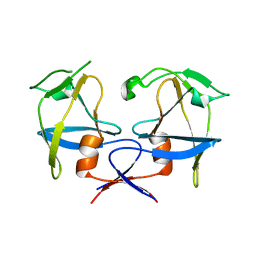 | | Crystal structure of dimeric M-PMV protease D26N mutant | | 分子名称: | Gag-Pro-Pol polyprotein | | 著者 | Wosicki, S, Gilski, M, Jaskolski, M, Zabranska, H, Pichova, I. | | 登録日 | 2019-06-19 | | 公開日 | 2019-10-16 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | Comparison of a retroviral protease in monomeric and dimeric states.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6S0R
 
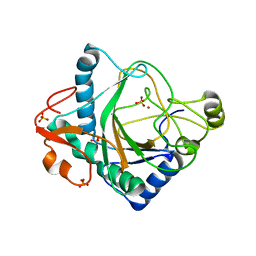 | | The crystal structure of kanamycin B dioxygenase (KanJ) from Streptomyces kanamyceticus complex with nickel, sulfate and chloride | | 分子名称: | CHLORIDE ION, Kanamycin B dioxygenase, NICKEL (II) ION, ... | | 著者 | Mrugala, B, Porebski, P.J, Niedzialkowska, E, Cymborowski, M.T, Minor, W, Borowski, T. | | 登録日 | 2019-06-18 | | 公開日 | 2020-07-08 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | A study on the structure, mechanism, and biochemistry of kanamycin B dioxygenase (KanJ)-an enzyme with a broad range of substrates.
Febs J., 288, 2021
|
|
6THE
 
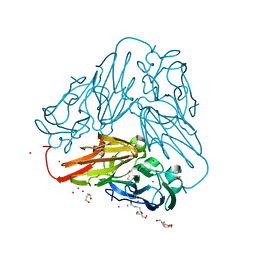 | | Crystal structure of core domain of four-domain heme-cupredoxin-Cu nitrite reductase from Bradyrhizobium sp. ORS 375 | | 分子名称: | CHLORIDE ION, COPPER (II) ION, Copper-containing nitrite reductase, ... | | 著者 | Sasaki, D, Watanabe, T.F, Eady, R.R, Garratt, R.C, Antonyuk, S.V, Hasnain, S.S. | | 登録日 | 2019-11-20 | | 公開日 | 2020-04-22 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.87 Å) | | 主引用文献 | Reverse protein engineering of a novel 4-domain copper nitrite reductase reveals functional regulation by protein-protein interaction.
Febs J., 288, 2021
|
|
5CFY
 
 | |
5DT1
 
 | |
6I0I
 
 | |
5FM1
 
 | | Structure of gamma-tubulin small complex based on a cryo-EM map, chemical cross-links, and a remotely related structure | | 分子名称: | SPINDLE POLE BODY COMPONENT 110, SPINDLE POLE BODY COMPONENT SPC97, SPINDLE POLE BODY COMPONENT SPC98, ... | | 著者 | Greenberg, C.H, Kollman, J, Zelter, A, Johnson, R, MacCoss, M.J, Davis, T.N, Agard, D.A, Sali, A. | | 登録日 | 2015-10-30 | | 公開日 | 2016-02-03 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (8 Å) | | 主引用文献 | Structure of Gamma-Tubulin Small Complex Based on a Cryo-Em Map, Chemical Cross-Links, and a Remotely Related Structure.
J.Struct.Biol., 194, 2016
|
|
6FHP
 
 | | DAIP in complex with a C-terminal fragment of thermolysin | | 分子名称: | Dispase autolysis-inducing protein, Thermolysin | | 著者 | Schmelz, S, Fiebig, D, Fuchsbauer, H.L, Blankenfeldt, W, Scrima, A. | | 登録日 | 2018-01-15 | | 公開日 | 2018-09-12 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.703 Å) | | 主引用文献 | Destructive twisting of neutral metalloproteases: the catalysis mechanism of the Dispase autolysis-inducing protein from Streptomyces mobaraensis DSM 40487.
FEBS J., 285, 2018
|
|
7DFS
 
 | | Crystal structure of a novel 4-O-alpha-L-rhamnosyl-beta-D-glucuronidase from Fusarium oxysporum 12S - Rha-GlcA complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-O-alpha-L-rhamnosyl-beta-D-glucuronidase, alpha-D-mannopyranose, ... | | 著者 | Kondo, T, Arakawa, T, Fushinobu, S, Sakamoto, T. | | 登録日 | 2020-11-09 | | 公開日 | 2021-03-17 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.49 Å) | | 主引用文献 | Biochemical and structural characterization of a novel 4-O-alpha-l-rhamnosyl-beta-d-glucuronidase from Fusarium oxysporum.
Febs J., 288, 2021
|
|
3J4Q
 
 | |
7DFQ
 
 | | Crystal Structure of a novel 4-O-alpha-L-rhamnosyl-beta-D-glucuronidase from Fusarium oxysporum 12S, ligand-free form | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-O-alpha-L-rhamnosyl-beta-D-glucuronidase | | 著者 | Kondo, T, Arakawa, T, Fushinobu, S, Sakamoto, T. | | 登録日 | 2020-11-09 | | 公開日 | 2021-03-17 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.51 Å) | | 主引用文献 | Biochemical and structural characterization of a novel 4-O-alpha-l-rhamnosyl-beta-d-glucuronidase from Fusarium oxysporum.
Febs J., 288, 2021
|
|
3J4U
 
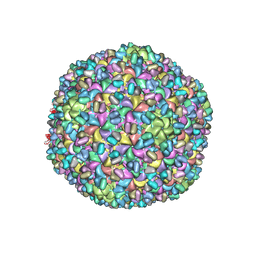 | | A new topology of the HK97-like fold revealed in Bordetella bacteriophage: non-covalent chainmail secured by jellyrolls | | 分子名称: | cementing protein, major capsid protein | | 著者 | Zhang, X, Guo, H, Jin, L, Czornyj, E, Hodes, A, Hui, W.H, Nieh, A.W, Miller, J.F, Zhou, Z.H. | | 登録日 | 2013-10-09 | | 公開日 | 2013-12-25 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | A new topology of the HK97-like fold revealed in Bordetella bacteriophage by cryoEM at 3.5 A resolution.
Elife, 2, 2013
|
|
3J6C
 
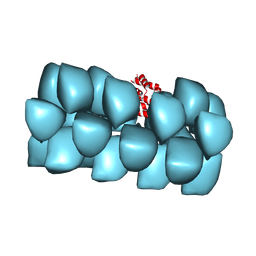 | | Cryo-EM structure of MAVS CARD filament | | 分子名称: | Mitochondrial antiviral-signaling protein | | 著者 | Xu, H, He, X, Zheng, H, Huang, L.J, Hou, F, Yu, Z, de la Cruz, M.J, Borkowski, B, Zhang, X, Chen, Z.J, Jiang, Q.-X. | | 登録日 | 2014-02-04 | | 公開日 | 2014-03-05 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (9.6 Å) | | 主引用文献 | Structural basis for the prion-like MAVS filaments in antiviral innate immunity.
Elife, 3, 2014
|
|
3J2Z
 
 | |
3J2W
 
 | |
3J4R
 
 | |
3J2Y
 
 | |
3J2X
 
 | |
3J30
 
 | |
7F3Y
 
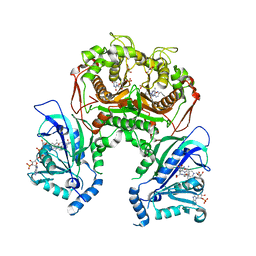 | | Wild-type Plasmodium falciparum dihydrofolate reductase-thymidylate synthase (PfDHFR-TS) complexed with methotrexate (MTX), NADPH and dUMP | | 分子名称: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, Bifunctional dihydrofolate reductase-thymidylate synthase, GLYCEROL, ... | | 著者 | Vanichtanankul, J, Tanramluk, D, Yuvaniyama, J, Yuthavong, Y. | | 登録日 | 2021-06-17 | | 公開日 | 2021-09-22 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.252 Å) | | 主引用文献 | MANORAA: A machine learning platform to guide protein-ligand design by anchors and influential distances.
Structure, 30, 2022
|
|
