8I6O
 
 | |
8I6R
 
 | | Cryo-EM structure of Pseudomonas aeruginosa FtsE(E163Q)X/EnvC complex with ATP in peptidisc | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Cell division ATP-binding protein FtsE, Cell division protein FtsX, ... | | 著者 | Xu, X, Li, J, Luo, M. | | 登録日 | 2023-01-29 | | 公開日 | 2023-06-07 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Mechanistic insights into the regulation of cell wall hydrolysis by FtsEX and EnvC at the bacterial division site.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8I6Q
 
 | |
8HVH
 
 | |
8HW4
 
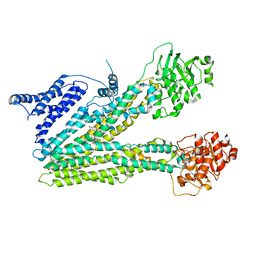 | | Cryo-EM structure of dehydroepiandrosterone sulfate-bound human ABC transporter ABCC3 in nanodiscs | | 分子名称: | 17-oxoandrost-5-en-3beta-yl hydrogen sulfate, ATP-binding cassette sub-family C member 3 | | 著者 | Wang, J, Wang, F.F, Chen, Y.X, Zhou, C.Z. | | 登録日 | 2022-12-28 | | 公開日 | 2023-06-07 | | 最終更新日 | 2023-10-11 | | 実験手法 | ELECTRON MICROSCOPY (3.52 Å) | | 主引用文献 | Placing steroid hormones within the human ABCC3 transporter reveals a compatible amphiphilic substrate-binding pocket.
Embo J., 42, 2023
|
|
8HW2
 
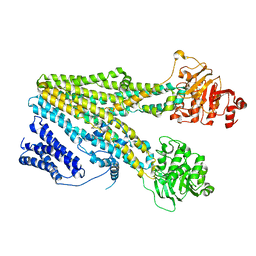 | | Cryo-EM structure of beta-estradiol 17-(beta-D-glucuronide)-bound human ABC transporter ABCC3 in nanodiscs | | 分子名称: | ATP-binding cassette sub-family C member 3, Estradiol-17beta-glucuronide | | 著者 | Wang, J, Wang, F.F, Chen, Y.X, Zhou, C.Z. | | 登録日 | 2022-12-28 | | 公開日 | 2023-06-07 | | 最終更新日 | 2023-10-11 | | 実験手法 | ELECTRON MICROSCOPY (3.65 Å) | | 主引用文献 | Placing steroid hormones within the human ABCC3 transporter reveals a compatible amphiphilic substrate-binding pocket.
Embo J., 42, 2023
|
|
8CAS
 
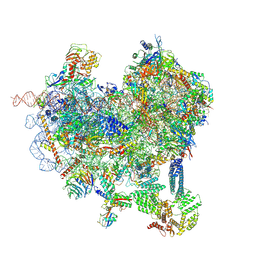 | | Cryo-EM structure of native Otu2-bound ubiquitinated 48S initiation complex (partial) | | 分子名称: | 18S ribosomal RNA, 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, ... | | 著者 | Ikeuchi, K, Buschauer, R, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | 登録日 | 2023-01-24 | | 公開日 | 2023-05-24 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Molecular basis for recognition and deubiquitination of 40S ribosomes by Otu2.
Nat Commun, 14, 2023
|
|
8CAH
 
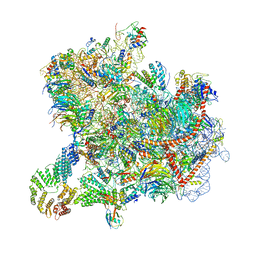 | | Cryo-EM structure of native Otu2-bound ubiquitinated 43S pre-initiation complex | | 分子名称: | 18S ribosomal RNA, 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, ... | | 著者 | Ikeuchi, K, Buschauer, R, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | 登録日 | 2023-01-24 | | 公開日 | 2023-05-24 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Molecular basis for recognition and deubiquitination of 40S ribosomes by Otu2.
Nat Commun, 14, 2023
|
|
8J3W
 
 | | Cryo-EM structure of aspirin-bound ABCC4 | | 分子名称: | 2-(ACETYLOXY)BENZOIC ACID, ATP-binding cassette sub-family C member 4 | | 著者 | Chen, Y, Wang, L, Hou, W.T, Zhou, C.Z, Chen, Y, Li, Q. | | 登録日 | 2023-04-18 | | 公開日 | 2023-05-24 | | 実験手法 | ELECTRON MICROSCOPY (3.07 Å) | | 主引用文献 | Cryo-EM structure ofABCC4
Nat Cardiovasc Res, 2023
|
|
8J3Z
 
 | | Cryo-EM structure of ATP-U46619-bound ABCC4 | | 分子名称: | (5Z)-7-{(1R,4S,5S,6R)-6-[(1E,3S)-3-hydroxyoct-1-en-1-yl]-2-oxabicyclo[2.2.1]hept-5-yl}hept-5-enoic acid, ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family C member 4, ... | | 著者 | Chen, Y, Wang, L, Hou, W.T, Zhou, C.Z, Chen, Y, Li, Q. | | 登録日 | 2023-04-18 | | 公開日 | 2023-05-24 | | 実験手法 | ELECTRON MICROSCOPY (3.17 Å) | | 主引用文献 | Cryo-EM structure ofABCC4
Nat Cardiovasc Res, 2023
|
|
8I4B
 
 | | Cryo-EM structure of apo-form ABCC4 | | 分子名称: | ATP-binding cassette sub-family C member 4 | | 著者 | Chen, Y, Wang, L, Hou, W.T, Zhou, C.Z, Chen, Y, Li, Q. | | 登録日 | 2023-01-19 | | 公開日 | 2023-05-24 | | 実験手法 | ELECTRON MICROSCOPY (3.13 Å) | | 主引用文献 | Cryo-EM structure ofABCC4
Nat Cardiovasc Res, 2023
|
|
8I4A
 
 | | Cryo-EM structure of dipyridamole-bound ABCC4 | | 分子名称: | 2-[[2-[bis(2-hydroxyethyl)amino]-4,8-di(piperidin-1-yl)pyrimido[5,4-d]pyrimidin-6-yl]-(2-hydroxyethyl)amino]ethanol, ATP-binding cassette sub-family C member 4 | | 著者 | Chen, Y, Wang, L, Hou, W.T, Zhou, C.Z, Chen, Y, Li, Q. | | 登録日 | 2023-01-19 | | 公開日 | 2023-05-24 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Cryo-EM structure ofABCC4
Nat Cardiovasc Res, 2023
|
|
8I4C
 
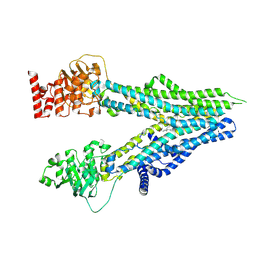 | | Cryo-EM structure of U46619-bound ABCC4 | | 分子名称: | (5Z)-7-{(1R,4S,5S,6R)-6-[(1E,3S)-3-hydroxyoct-1-en-1-yl]-2-oxabicyclo[2.2.1]hept-5-yl}hept-5-enoic acid, ATP-binding cassette sub-family C member 4 | | 著者 | Chen, Y, Wang, L, Hou, W.T, Zhou, C.Z, Chen, Y, Li, Q. | | 登録日 | 2023-01-19 | | 公開日 | 2023-05-24 | | 実験手法 | ELECTRON MICROSCOPY (3.08 Å) | | 主引用文献 | Cryo-EM structure ofABCC4
Nat Cardiovasc Res, 2023
|
|
7Y48
 
 | |
7ZK4
 
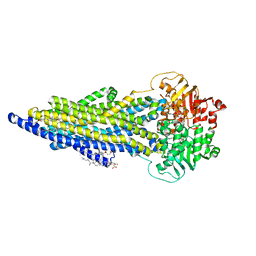 | | The ABCB1 L335C mutant (mABCB1) in the outward facing state | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent translocase ABCB1, CHOLESTEROL HEMISUCCINATE, ... | | 著者 | Parey, K, Januliene, D, Gewering, T, Moeller, A. | | 登録日 | 2022-04-12 | | 公開日 | 2023-04-26 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (2.6 Å) | | 主引用文献 | Tracing the substrate translocation mechanism in P-glycoprotein.
Elife, 12, 2024
|
|
7ZK6
 
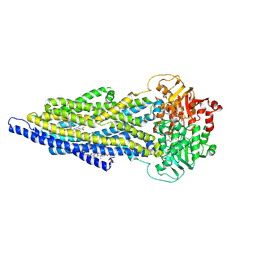 | | ABCB1 L335C mutant (mABCB1) in the outward facing state bound to 2 molecules of AAC | | 分子名称: | (4S,11S,18S)-4-[[(2,4-dinitrophenyl)disulfanyl]methyl]-11,18-dimethyl-6,13,20-trithia-3,10,17,22,23,24-hexazatetracyclo[17.2.1.1^{5,8}.1^{12,15}]tetracosa-1(21),5(24),7,12(23),14,19(22)-hexaene-2,9,16-trione, (4~{S},11~{S},18~{S})-4,11-dimethyl-18-(sulfanylmethyl)-6,13,20-trithia-3,10,17,22,23,24-hexazatetracyclo[17.2.1.1^{5,8}.1^{12,15}]tetracosa-1(21),5(24),7,12(23),14,19(22)-hexaene-2,9,16-trione, ADENOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Parey, K, Januliene, D, Gewering, T, Moeller, A. | | 登録日 | 2022-04-12 | | 公開日 | 2023-04-26 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Tracing the substrate translocation mechanism in P-glycoprotein.
Elife, 12, 2024
|
|
7ZKA
 
 | | ABCB1 V978C mutant (mABCB1) in the outward facing state bound to AAC | | 分子名称: | (4~{S},11~{S},18~{S})-4,11-dimethyl-18-(sulfanylmethyl)-6,13,20-trithia-3,10,17,22,23,24-hexazatetracyclo[17.2.1.1^{5,8}.1^{12,15}]tetracosa-1(21),5(24),7,12(23),14,19(22)-hexaene-2,9,16-trione, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent translocase ABCB1, ... | | 著者 | Parey, K, Januliene, D, Gewering, T, Urbatsch, I, Zhang, Q, Moeller, A. | | 登録日 | 2022-04-12 | | 公開日 | 2023-04-26 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Tracing the substrate translocation mechanism in P-glycoprotein.
Elife, 12, 2024
|
|
7ZK9
 
 | | ABCB1 L971C mutant (mABCB1) in the inward facing state | | 分子名称: | (4~{S},11~{S},18~{S})-4,11-dimethyl-18-(sulfanylmethyl)-6,13,20-trithia-3,10,17,22,23,24-hexazatetracyclo[17.2.1.1^{5,8}.1^{12,15}]tetracosa-1(21),5(24),7,12(23),14,19(22)-hexaene-2,9,16-trione, ATP-dependent translocase ABCB1 | | 著者 | Parey, K, Januliene, D, Gewering, T, Zhang, Q, Moeller, A. | | 登録日 | 2022-04-12 | | 公開日 | 2023-04-26 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | Tracing the substrate translocation mechanism in P-glycoprotein.
Elife, 12, 2024
|
|
7ZK5
 
 | | ABCB1 L335C mutant (mABCB1) in the outward facing state bound to AAC | | 分子名称: | (4~{S},11~{S},18~{S})-4,11-dimethyl-18-(sulfanylmethyl)-6,13,20-trithia-3,10,17,22,23,24-hexazatetracyclo[17.2.1.1^{5,8}.1^{12,15}]tetracosa-1(21),5(24),7,12(23),14,19(22)-hexaene-2,9,16-trione, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent translocase ABCB1, ... | | 著者 | Parey, K, Januliene, D, Gewering, T, Moeller, A. | | 登録日 | 2022-04-12 | | 公開日 | 2023-04-26 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (2.6 Å) | | 主引用文献 | Tracing the substrate translocation mechanism in P-glycoprotein.
Elife, 12, 2024
|
|
7ZKB
 
 | | ABCB1 V978C mutant (mABCB1) in the inward facing state | | 分子名称: | (4S,11S,18S)-4-[[(2,4-dinitrophenyl)disulfanyl]methyl]-11,18-dimethyl-6,13,20-trithia-3,10,17,22,23,24-hexazatetracyclo[17.2.1.1^{5,8}.1^{12,15}]tetracosa-1(21),5(24),7,12(23),14,19(22)-hexaene-2,9,16-trione, (4~{S},11~{S},18~{S})-4,11-dimethyl-18-(sulfanylmethyl)-6,13,20-trithia-3,10,17,22,23,24-hexazatetracyclo[17.2.1.1^{5,8}.1^{12,15}]tetracosa-1(21),5(24),7,12(23),14,19(22)-hexaene-2,9,16-trione, ATP-dependent translocase ABCB1 | | 著者 | Parey, K, Januliene, D, Gewering, T, Moeller, A. | | 登録日 | 2022-04-12 | | 公開日 | 2023-04-26 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (4.7 Å) | | 主引用文献 | Tracing the substrate translocation mechanism in P-glycoprotein.
Elife, 12, 2024
|
|
7ZK8
 
 | | ABCB1 L971C mutant (mABCB1) in the outward facing state bound to AAC | | 分子名称: | (4~{S},11~{S},18~{S})-4,11-dimethyl-18-(sulfanylmethyl)-6,13,20-trithia-3,10,17,22,23,24-hexazatetracyclo[17.2.1.1^{5,8}.1^{12,15}]tetracosa-1(21),5(24),7,12(23),14,19(22)-hexaene-2,9,16-trione, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent translocase ABCB1, ... | | 著者 | Parey, K, Januliene, D, Gewering, T, Moeller, A. | | 登録日 | 2022-04-12 | | 公開日 | 2023-04-26 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Tracing the substrate translocation mechanism in P-glycoprotein.
Elife, 12, 2024
|
|
7ZDF
 
 | |
7ZDU
 
 | |
7ZDC
 
 | |
7ZDA
 
 | |
