6PG4
 
 | |
5KKO
 
 | | A 1.55A X-Ray Structure from Vibrio cholerae O1 biovar El Tor of a Hypothetical Protein | | 分子名称: | Uncharacterised protein | | 著者 | Brunzelle, J.S, Wawrzak, Z, Skarina, T, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2016-06-22 | | 公開日 | 2016-09-21 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | A 1.55A X-Ray Structure from Vibrio cholerae O1 biovar El Tor of a Hypothetical Protein
To Be Published
|
|
6PG9
 
 | |
5W0A
 
 | | Crystal structure of Trichoderma harzianum endoglucanase I | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glucanase, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Godoy, A.S, Pellegrini, V.O.A, Sonoda, M.T, Kadowaki, M.A, Nascimento, A.S, Polikarpov, I. | | 登録日 | 2017-05-30 | | 公開日 | 2018-05-30 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.898 Å) | | 主引用文献 | Structure and dynamics of Trichoderma harzianum Cel7B suggest molecular architecture adaptations required for a wide spectrum of activities on plant cell wall polysaccharides.
Biochim Biophys Acta Gen Subj, 1863, 2019
|
|
6N3G
 
 | | Crystal structure of histone lysine methyltransferase SmyD2 in complex with polyethylene glycol | | 分子名称: | DODECAETHYLENE GLYCOL, ETHANOL, N-lysine methyltransferase SMYD2, ... | | 著者 | Perry, E, Spellmon, N, Brunzelle, J, Yang, Z. | | 登録日 | 2018-11-15 | | 公開日 | 2019-11-20 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.43 Å) | | 主引用文献 | Crystal structure of histone lysine methyltransferase SmyD2 in complex with polyethylene glycol
To be Published
|
|
6G0B
 
 | |
5W0U
 
 | | Crystal structure of MBP fused activation-induced cytidine deaminase (AID) in complex with dCMP | | 分子名称: | 2'-DEOXYCYTIDINE-5'-MONOPHOSPHATE, CALCIUM ION, DNA (5'-D(*CP*TP*GP*GP*CP*CP*TP*TP*GP*AP*AP*C)-3'), ... | | 著者 | Qiao, Q, Wang, L, Wu, H. | | 登録日 | 2017-05-31 | | 公開日 | 2017-08-16 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | AID Recognizes Structured DNA for Class Switch Recombination.
Mol. Cell, 67, 2017
|
|
6N3S
 
 | | Crystal structure of apo-cruzain | | 分子名称: | 1,2-ETHANEDIOL, Cruzipain, PHOSPHATE ION | | 著者 | Silva, E.B, Dall, E, Rodrigues, F.T.G, Ferreira, R.S, Brandstetter, H. | | 登録日 | 2018-11-16 | | 公開日 | 2019-05-29 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.193 Å) | | 主引用文献 | Cruzain structures: apocruzain and cruzain bound to S-methyl thiomethanesulfonate and implications for drug design.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
5E02
 
 | |
7CK4
 
 | |
6PHL
 
 | |
6N7Q
 
 | |
5KKW
 
 | |
5DZX
 
 | | Protocadherin beta 6 extracellular cadherin domains 1-4 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Protocadherin beta 6, ... | | 著者 | Goodman, K.M, Mannepalli, S, Bahna, F, Honig, B, Shapiro, L. | | 登録日 | 2015-09-26 | | 公開日 | 2016-05-04 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.879 Å) | | 主引用文献 | Structural Basis of Diverse Homophilic Recognition by Clustered alpha- and beta-Protocadherins.
Neuron, 90, 2016
|
|
6GAS
 
 | |
6UTP
 
 | | LarE, a sulfur transferase involved in synthesis of the cofactor for lactate racemase in complex with cobalt | | 分子名称: | ATP-dependent sacrificial sulfur transferase LarE, COBALT (II) ION, PHOSPHATE ION, ... | | 著者 | Fellner, M, Huizenga, K, Hausinger, R.P, Hu, J. | | 登録日 | 2019-10-29 | | 公開日 | 2019-11-06 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (3.55 Å) | | 主引用文献 | Crystallographic characterization of a tri-Asp metal-binding site at the three-fold symmetry axis of LarE.
Sci Rep, 10, 2020
|
|
6PHQ
 
 | |
6G0X
 
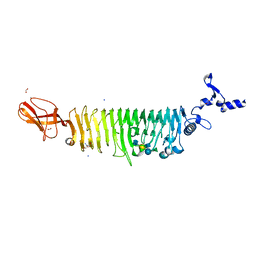 | | TAILSPIKE PROTEIN OF E. COLI BACTERIOPHAGE HK620 IN COMPLEX WITH PENTASACCHARIDE | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FORMIC ACID, SODIUM ION, ... | | 著者 | Gohlke, U, Broeker, N.K, Seckler, R, Barbirz, S. | | 登録日 | 2018-03-20 | | 公開日 | 2018-04-04 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.41 Å) | | 主引用文献 | Solvent Networks Tune Thermodynamics of Oligosaccharide Complex Formation in an Extended Protein Binding Site.
J. Am. Chem. Soc., 140, 2018
|
|
6YHM
 
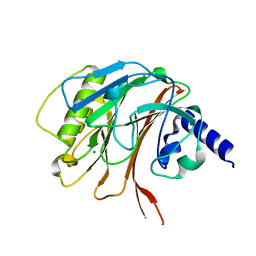 | | Crystal structure of the C-terminal domain of CNFy from Yersinia pseudotuberculosis | | 分子名称: | Cytotoxic necrotizing factor, MAGNESIUM ION | | 著者 | Lukat, P, Gazdag, E.M, Heidler, T.V, Blankenfeldt, W. | | 登録日 | 2020-03-30 | | 公開日 | 2020-12-30 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.13 Å) | | 主引用文献 | Crystal structure of bacterial cytotoxic necrotizing factor CNF Y reveals molecular building blocks for intoxication.
Embo J., 40, 2021
|
|
5KLF
 
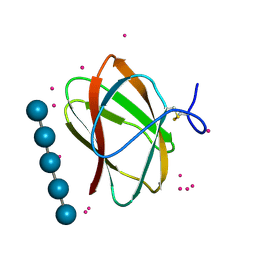 | | Structure of CBM_E1, a novel carbohydrate-binding module found by sugar cane soil metagenome, complexed with cellopentaose and gadolinium ion | | 分子名称: | Carbohydrate binding module E1, GADOLINIUM ATOM, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | 著者 | Liberato, M.V, Campos, B.M, Zeri, A.C.M, Squina, F.M. | | 登録日 | 2016-06-24 | | 公開日 | 2016-09-21 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.801 Å) | | 主引用文献 | A Novel Carbohydrate-binding Module from Sugar Cane Soil Metagenome Featuring Unique Structural and Carbohydrate Affinity Properties.
J.Biol.Chem., 291, 2016
|
|
5HW5
 
 | |
6VER
 
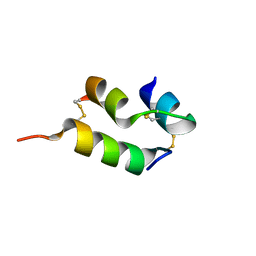 | | Human insulin analog: [GluB10,TyrB20]-DOI | | 分子名称: | Insulin A chain, Insulin B chain | | 著者 | Menting, J.G, Chou, D.H.-C, Lawrence, M.C, Xiong, X. | | 登録日 | 2020-01-02 | | 公開日 | 2020-11-11 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.047 Å) | | 主引用文献 | Mini-Ins: A minimal, bioactive insulin analog with alternative binding modes
not published
|
|
6G1F
 
 | |
5KLR
 
 | | Prototypical P4[R]cNLS | | 分子名称: | Importin subunit alpha-1, Prototypical P4[R]cNLS | | 著者 | Smith, K.M, Forwood, J.K. | | 登録日 | 2016-06-25 | | 公開日 | 2016-07-27 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Contribution of the residue at position 4 within classical nuclear localization signals to modulating interaction with importins and nuclear targeting.
Biochim Biophys Acta Mol Cell Res, 1865, 2018
|
|
6PI1
 
 | |
