6FTZ
 
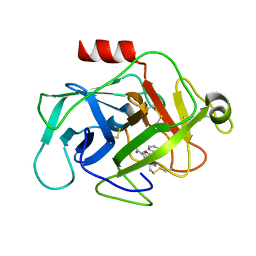 | | COMPLEMENT FACTOR D COMPLEXED WITH COMPOUND 6 | | 分子名称: | Complement factor D, ~{N}4-[3-(aminomethyl)phenyl]-1~{H}-indole-2,4-dicarboxamide | | 著者 | Ostermann, N. | | 登録日 | 2018-02-26 | | 公開日 | 2018-06-06 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.67 Å) | | 主引用文献 | Discovery and Design of First Benzylamine-Based Ligands Binding to an Unlocked Conformation of the Complement Factor D.
ACS Med Chem Lett, 9, 2018
|
|
6FUH
 
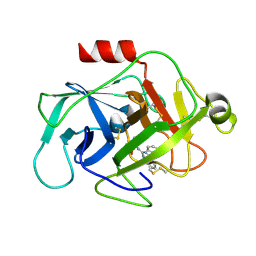 | | Complement factor D in complex with the inhibitor (4-((3-(aminomethyl)phenyl)amino)quinazolin-2-yl)-L-valine | | 分子名称: | (2~{S})-2-[[4-[[3-(aminomethyl)phenyl]amino]quinazolin-2-yl]amino]-3-methyl-butanoic acid, Complement factor D | | 著者 | Mac Sweeney, A, Ostermann, N, Vulpetti, A, Maibaum, J, Erbel, P, Lorthiois, E, Yoon, T, Randl, S, Ruedisser, S. | | 登録日 | 2018-02-27 | | 公開日 | 2018-06-06 | | 実験手法 | X-RAY DIFFRACTION (1.37 Å) | | 主引用文献 | Discovery and Design of First Benzylamine-Based Ligands Binding to an Unlocked Conformation of the Complement Factor D.
ACS Med Chem Lett, 9, 2018
|
|
7A5S
 
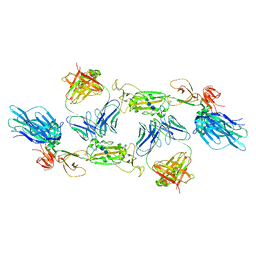 | | Complex of SARS-CoV-2 spike and CR3022 Fab (Homogeneous Refinement) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CR3022 Fab Heavy Chain, CR3022 Fab Light Chain, ... | | 著者 | Wrobel, A.G, Benton, D.J, Rosenthal, P.B, Gamblin, S.J. | | 登録日 | 2020-08-21 | | 公開日 | 2020-09-16 | | 最終更新日 | 2020-11-04 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Antibody-mediated disruption of the SARS-CoV-2 spike glycoprotein.
Nat Commun, 11, 2020
|
|
7A5R
 
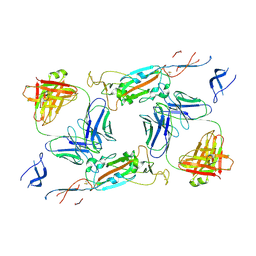 | | Complex of SARS-CoV-2 spike and CR3022 Fab (Non-Uniform Refinement) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CR3022 Fab Heavy Chain, CR3022 Fab Light Chain, ... | | 著者 | Wrobel, A.G, Benton, D.J, Rosenthal, P.B, Gamblin, S.J. | | 登録日 | 2020-08-21 | | 公開日 | 2020-09-16 | | 最終更新日 | 2020-11-04 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Antibody-mediated disruption of the SARS-CoV-2 spike glycoprotein.
Nat Commun, 11, 2020
|
|
6FQO
 
 | | Crystal structure of CREBBP bromodomain complexd with DT29 | | 分子名称: | 1,2-ETHANEDIOL, CREB-binding protein, ~{N}-[3-(2,5-dimethyl-3-oxidanylidene-1,2-oxazol-4-yl)-5-(5-ethanoyl-2-ethoxy-phenyl)phenyl]furan-2-carboxamide | | 著者 | Zhu, J, Caflisch, A. | | 登録日 | 2018-02-14 | | 公開日 | 2018-08-29 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Binding Motifs in the CBP Bromodomain: An Analysis of 20 Crystal Structures of Complexes with Small Molecules.
ACS Med Chem Lett, 9, 2018
|
|
6FQU
 
 | | Crystal structure of CREBBP bromodomain complexd with DR09 | | 分子名称: | 1-[3-[3-[3,3-bis(fluoranyl)piperidin-1-yl]phenyl]-4-ethoxy-phenyl]ethanone, CREB-binding protein | | 著者 | Zhu, J, Caflisch, A. | | 登録日 | 2018-02-14 | | 公開日 | 2018-08-29 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.43 Å) | | 主引用文献 | Binding Motifs in the CBP Bromodomain: An Analysis of 20 Crystal Structures of Complexes with Small Molecules.
ACS Med Chem Lett, 9, 2018
|
|
6FUI
 
 | | Complement factor D in complex with the inhibitor 3-((3-((3-(aminomethyl)phenyl)amino)-1H-pyrazolo[3,4-d]pyrimidin-4-yl)amino)phenol | | 分子名称: | (1~{R},2~{S})-2-[[4-[[3-(aminomethyl)phenyl]amino]quinazolin-2-yl]amino]cyclohexane-1-carboxylic acid, Complement factor D | | 著者 | Mac Sweeney, A, Ostermann, N, Vulpetti, A, Maibaum, J, Erbel, P, Lorthiois, E, Yoon, T, Randl, S, Ruedisser, S. | | 登録日 | 2018-02-27 | | 公開日 | 2018-06-06 | | 実験手法 | X-RAY DIFFRACTION (1.38 Å) | | 主引用文献 | Discovery and Design of First Benzylamine-Based Ligands Binding to an Unlocked Conformation of the Complement Factor D.
ACS Med Chem Lett, 9, 2018
|
|
6T3F
 
 | | Crystal structure Nipah virus fusion glycoprotein in complex with a neutralising Fab fragment | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab66 heavy chain, ... | | 著者 | Avanzato, V.A, Pryce, R, Walter, T.S, Bowden, T.A. | | 登録日 | 2019-10-10 | | 公開日 | 2019-11-27 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | A structural basis for antibody-mediated neutralization of Nipah virus reveals a site of vulnerability at the fusion glycoprotein apex.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
2BDA
 
 | | Porcine pancreatic elastase complexed with N-acetyl-NPI and Ala-Ala at pH 5.0 | | 分子名称: | ALANINE, CALCIUM ION, Elastase-1, ... | | 著者 | Liu, B, Schofield, C.J, Wilmouth, R.C. | | 登録日 | 2005-10-20 | | 公開日 | 2006-05-30 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural analyses on intermediates in serine protease catalysis
J.Biol.Chem., 281, 2006
|
|
2BDC
 
 | |
2BDB
 
 | | Porcine pancreatic elastase complexed with Asn-Pro-Ile and Ala-Ala at pH 5.0 | | 分子名称: | ALANINE, CALCIUM ION, Elastase-1, ... | | 著者 | Liu, B, Schofield, C.J, Wilmouth, R.C. | | 登録日 | 2005-10-20 | | 公開日 | 2006-05-30 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural analyses on intermediates in serine protease catalysis
J.Biol.Chem., 281, 2006
|
|
4N1Z
 
 | | Crystal Structure of Human Farnesyl Diphosphate Synthase in Complex with BPH-1222 | | 分子名称: | Farnesyl pyrophosphate synthase, MAGNESIUM ION, PHOSPHATE ION, ... | | 著者 | Liu, Y.L, Xia, Y, Zhang, Y, Verma, I, Oldfield, E. | | 登録日 | 2013-10-04 | | 公開日 | 2014-12-10 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | A combination therapy for KRAS-driven lung adenocarcinomas using lipophilic bisphosphonates and rapamycin.
Sci Transl Med, 6, 2014
|
|
2BD2
 
 | | Porcine pancreatic elastase complexed with beta-casomorphin-7 and Arg-Phe at pH 5.0 | | 分子名称: | ARGININE, CALCIUM ION, Elastase-1, ... | | 著者 | Liu, B, Schofield, C.J, Wilmouth, R.C. | | 登録日 | 2005-10-20 | | 公開日 | 2006-05-30 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural analyses on intermediates in serine protease catalysis
J.Biol.Chem., 281, 2006
|
|
6FUJ
 
 | | Complement factor D in complex with the inhibitor N-(3'-(aminomethyl)-[1,1'-biphenyl]-3-yl)-3-methylbutanamide | | 分子名称: | Complement factor D, ~{N}-[3-[3-(aminomethyl)phenyl]phenyl]-3-methyl-butanamide | | 著者 | Mac Sweeney, A, Ostermann, N, Vulpetti, A, Maibaum, J, Erbel, P, Lorthiois, E, Yoon, T, Randl, S, Ruedisser, S. | | 登録日 | 2018-02-27 | | 公開日 | 2018-06-06 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Discovery and Design of First Benzylamine-Based Ligands Binding to an Unlocked Conformation of the Complement Factor D.
ACS Med Chem Lett, 9, 2018
|
|
2BD7
 
 | | Porcine pancreatic elastase complexed with beta-casomorphin-7 and Arg-Phe at pH 5.0 (50 min soak) | | 分子名称: | ARGININE, CALCIUM ION, Elastase-1, ... | | 著者 | Liu, B, Schofield, C.J, Wilmouth, R.C. | | 登録日 | 2005-10-20 | | 公開日 | 2006-05-30 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural analyses on intermediates in serine protease catalysis
J.Biol.Chem., 281, 2006
|
|
2BD3
 
 | | Porcine pancreatic elastase complexed with beta-casomorphin-7 and Lys-Ala-NH2 at pH 5.0 | | 分子名称: | ALANINE, CALCIUM ION, Elastase-1, ... | | 著者 | Liu, B, Schofield, C.J, Wilmouth, R.C. | | 登録日 | 2005-10-20 | | 公開日 | 2006-05-30 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural analyses on intermediates in serine protease catalysis
J.Biol.Chem., 281, 2006
|
|
6FTY
 
 | | COMPLEMENT FACTOR D COMPLEXED WITH COMPOUND 5 | | 分子名称: | 4-[[(5~{S},7~{R})-3-azanyl-1-adamantyl]carbonylamino]-1~{H}-indole-2-carboxamide, Complement factor D, GLYCEROL | | 著者 | Ostermann, N. | | 登録日 | 2018-02-26 | | 公開日 | 2018-06-06 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.67 Å) | | 主引用文献 | Discovery and Design of First Benzylamine-Based Ligands Binding to an Unlocked Conformation of the Complement Factor D.
ACS Med Chem Lett, 9, 2018
|
|
3N84
 
 | |
2WKS
 
 | | Structure of Helicobacter pylori Type II Dehydroquinase with a new carbasugar-thiophene inhibitor. | | 分子名称: | (1R,4S,5R)-1,4,5-trihydroxy-3-[(5-methyl-1-benzothiophen-2-yl)methoxy]cyclohex-2-ene-1-carboxylic acid, 3-DEHYDROQUINATE DEHYDRATASE | | 著者 | Otero, J.M, Guardado-Calvo, P, Llamas-Saiz, A.L, Prazeres, V.F.V, Tizon, L, Castedo, L, Lamb, H, Hawkins, A.R, Gonzalez-Bello, C, van Raaij, M.J. | | 登録日 | 2009-06-17 | | 公開日 | 2009-11-24 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.95 Å) | | 主引用文献 | Synthesis and biological evaluation of new nanomolar competitive inhibitors of Helicobacter pylori type II dehydroquinase. Structural details of the role of the aromatic moieties with essential residues.
J. Med. Chem., 53, 2010
|
|
6EB1
 
 | | HIV-1 Integrase Catalytic Core Domain Complexed with Allosteric Inhibitor (2S)-tert-butoxy[3-(3,4-dihydro-2H-1-benzopyran-6-yl)-1-phenylisoquinolin-4-yl]acetic acid | | 分子名称: | (2S)-tert-butoxy[3-(3,4-dihydro-2H-1-benzopyran-6-yl)-1-phenylisoquinolin-4-yl]acetic acid, Integrase | | 著者 | Lindenberger, J.J, Kobe, M, Kvaratskhelia, M. | | 登録日 | 2018-08-03 | | 公開日 | 2019-03-06 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | An Isoquinoline Scaffold as a Novel Class of Allosteric HIV-1 Integrase Inhibitors.
ACS Med Chem Lett, 10, 2019
|
|
1C8S
 
 | | BACTERIORHODOPSIN D96N LATE M STATE INTERMEDIATE | | 分子名称: | 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, 2,10,23-TRIMETHYL-TETRACOSANE, BACTERIORHODOPSIN ("M" STATE INTERMEDIATE), ... | | 著者 | Luecke, H. | | 登録日 | 1999-07-29 | | 公開日 | 1999-10-20 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural changes in bacteriorhodopsin during ion transport at 2 angstrom resolution.
Science, 286, 1999
|
|
4MAN
 
 | | Bcl_2-Navitoclax Analog (with Indole) Complex | | 分子名称: | 4-[4-({4'-chloro-3-[2-(dimethylamino)ethoxy]biphenyl-2-yl}methyl)piperazin-1-yl]-2-(1H-indol-5-yloxy)-N-({3-nitro-4-[(tetrahydro-2H-pyran-4-ylmethyl)amino]phenyl}sulfonyl)benzamide, Apoptosis regulator Bcl-2 | | 著者 | Park, C.H. | | 登録日 | 2013-08-16 | | 公開日 | 2014-01-29 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.07 Å) | | 主引用文献 | ABT-199, a potent and selective BCL-2 inhibitor, achieves antitumor activity while sparing platelets.
NAT.MED. (N.Y.), 19, 2013
|
|
6TKH
 
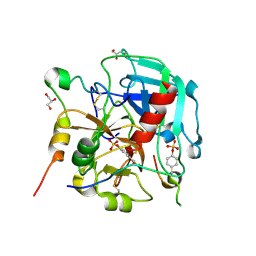 | | Tsetse thrombin inhibitor in complex with human alpha-thrombin - orthorhombic form at 7keV | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, SODIUM ION, ... | | 著者 | Calisto, B.M, Ripoll-Rozada, J, de Sanctis, D, Pereira, P.J.B. | | 登録日 | 2019-11-28 | | 公開日 | 2020-11-04 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Sulfotyrosine-Mediated Recognition of Human Thrombin by a Tsetse Fly Anticoagulant Mimics Physiological Substrates.
Cell Chem Biol, 28, 2021
|
|
4LVT
 
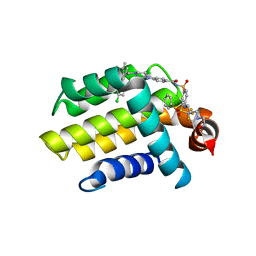 | | Bcl_2-Navitoclax (ABT-263) Complex | | 分子名称: | 4-(4-{[2-(4-chlorophenyl)-5,5-dimethylcyclohex-1-en-1-yl]methyl}piperazin-1-yl)-N-[(4-{[(2R)-4-(morpholin-4-yl)-1-(phenylsulfanyl)butan-2-yl]amino}-3-[(trifluoromethyl)sulfonyl]phenyl)sulfonyl]benzamide, Apoptosis regulator Bcl-2 | | 著者 | Park, C.H. | | 登録日 | 2013-07-26 | | 公開日 | 2013-08-14 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | ABT-199, a potent and selective BCL-2 inhibitor, achieves antitumor activity while sparing platelets.
NAT.MED. (N.Y.), 19, 2013
|
|
2BCG
 
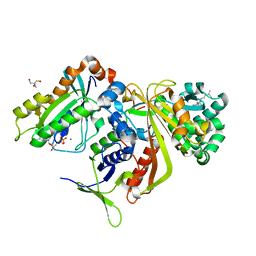 | | Structure of doubly prenylated Ypt1:GDI complex | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GERAN-8-YL GERAN, GTP-binding protein YPT1, ... | | 著者 | Pylypenko, O, Rak, A, Alexandrov, K. | | 登録日 | 2005-10-19 | | 公開日 | 2006-01-17 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.48 Å) | | 主引用文献 | Structure of doubly prenylated Ypt1:GDI complex and the mechanism of GDI-mediated Rab recycling
Embo J., 25, 2006
|
|
