6FNX
 
 | |
7AM5
 
 | |
3J7N
 
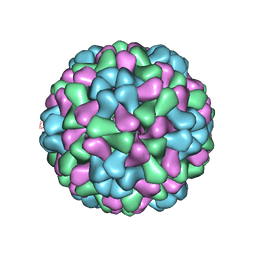 | | Virus model of brome mosaic virus (second half data set) | | 分子名称: | Capsid protein | | 著者 | Wang, Z, Hryc, C, Bammes, B, Afonine, P.V, Jakana, J, Chen, D.H, Liu, X, Baker, M.L, Kao, C, Ludtke, S.J, Schmid, M.F, Adams, P.D, Chiu, W. | | 登録日 | 2014-07-18 | | 公開日 | 2014-09-10 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | An atomic model of brome mosaic virus using direct electron detection and real-space optimization.
Nat Commun, 5, 2014
|
|
3J7L
 
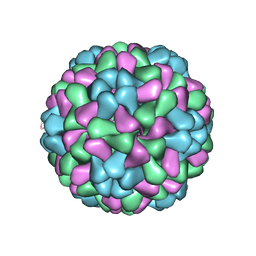 | | Full virus map of brome mosaic virus | | 分子名称: | Capsid protein | | 著者 | Wang, Z, Hryc, C, Bammes, B, Afonine, P.V, Jakana, J, Chen, D.H, Liu, X, Baker, M.L, Kao, C, Ludtke, S.J, Schmid, M.F, Adams, P.D, Chiu, W. | | 登録日 | 2014-07-18 | | 公開日 | 2014-09-10 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | An atomic model of brome mosaic virus using direct electron detection and real-space optimization.
Nat Commun, 5, 2014
|
|
5RG8
 
 | | Crystal Structure of Kemp Eliminase HG3.17 in unbound state, 277K | | 分子名称: | ACETATE ION, Kemp Eliminase HG3 | | 著者 | Broom, A, Rakotoharisoa, R.V, Thompson, M.C, Fraser, J.S, Chica, R.A. | | 登録日 | 2020-03-19 | | 公開日 | 2020-07-22 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.73 Å) | | 主引用文献 | Ensemble-based enzyme design can recapitulate the effects of laboratory directed evolution in silico.
Nat Commun, 11, 2020
|
|
5RG6
 
 | | Crystal Structure of Kemp Eliminase HG3.7 in unbound state, 277K | | 分子名称: | Kemp Eliminase HG3, SULFATE ION | | 著者 | Broom, A, Rakotoharisoa, R.V, Thompson, M.C, Fraser, J.S, Chica, R.A. | | 登録日 | 2020-03-19 | | 公開日 | 2020-07-22 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Ensemble-based enzyme design can recapitulate the effects of laboratory directed evolution in silico.
Nat Commun, 11, 2020
|
|
5RG5
 
 | | Crystal Structure of Kemp Eliminase HG3.3b in unbound state, 277K | | 分子名称: | ACETATE ION, Kemp Eliminase HG3, SULFATE ION | | 著者 | Broom, A, Rakotoharisoa, R.V, Thompson, M.C, Fraser, J.S, Chica, R.A. | | 登録日 | 2020-03-19 | | 公開日 | 2020-07-22 | | 最終更新日 | 2021-05-12 | | 実験手法 | X-RAY DIFFRACTION (1.62 Å) | | 主引用文献 | Ensemble-based enzyme design can recapitulate the effects of laboratory directed evolution in silico.
Nat Commun, 11, 2020
|
|
5RG7
 
 | | Crystal Structure of Kemp Eliminase HG3.14 in unbound state, 277K | | 分子名称: | Kemp Eliminase HG3, SULFATE ION | | 著者 | Broom, A, Rakotoharisoa, R.V, Thompson, M.C, Fraser, J.S, Chica, R.A. | | 登録日 | 2020-03-19 | | 公開日 | 2020-07-22 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.47 Å) | | 主引用文献 | Ensemble-based enzyme design can recapitulate the effects of laboratory directed evolution in silico.
Nat Commun, 11, 2020
|
|
5RGE
 
 | | Crystal Structure of Kemp Eliminase HG3.17 with bound transition state analog, 277K | | 分子名称: | 6-NITROBENZOTRIAZOLE, Kemp Eliminase HG3 | | 著者 | Broom, A, Rakotoharisoa, R.V, Thompson, M.C, Fraser, J.S, Chica, R.A. | | 登録日 | 2020-03-19 | | 公開日 | 2020-07-22 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | Ensemble-based enzyme design can recapitulate the effects of laboratory directed evolution in silico.
Nat Commun, 11, 2020
|
|
5RGB
 
 | | Crystal Structure of Kemp Eliminase HG3.3b with bound transition state analogue, 277K | | 分子名称: | 6-NITROBENZOTRIAZOLE, Kemp Eliminase HG3.3b, SULFATE ION | | 著者 | Broom, A, Rakotoharisoa, R.V, Thompson, M.C, Fraser, J.S, Chica, R.A. | | 登録日 | 2020-03-19 | | 公開日 | 2020-07-22 | | 最終更新日 | 2021-05-12 | | 実験手法 | X-RAY DIFFRACTION (1.42 Å) | | 主引用文献 | Ensemble-based enzyme design can recapitulate the effects of laboratory directed evolution in silico.
Nat Commun, 11, 2020
|
|
5RGD
 
 | | Crystal Structure of Kemp Eliminase HG3.14 with bound transition state analogue, 277K | | 分子名称: | 6-NITROBENZOTRIAZOLE, Kemp Eliminase HG3.14, SULFATE ION | | 著者 | Broom, A, Rakotoharisoa, R.V, Thompson, M.C, Fraser, J.S, Chica, R.A. | | 登録日 | 2020-03-19 | | 公開日 | 2020-07-22 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Ensemble-based enzyme design can recapitulate the effects of laboratory directed evolution in silico.
Nat Commun, 11, 2020
|
|
5RG4
 
 | | Crystal Structure of Kemp Eliminase HG3 in unbound state, 277K | | 分子名称: | ACETATE ION, Kemp Eliminase HG3, SULFATE ION | | 著者 | Broom, A, Rakotoharisoa, R.V, Thompson, M.C, Fraser, J.S, Chica, R.A. | | 登録日 | 2020-03-19 | | 公開日 | 2020-07-22 | | 最終更新日 | 2021-05-12 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Ensemble-based enzyme design can recapitulate the effects of laboratory directed evolution in silico.
Nat Commun, 11, 2020
|
|
5RGC
 
 | | Crystal Structure of Kemp Eliminase HG3.7 with bound transition state analogue, 277K | | 分子名称: | 6-NITROBENZOTRIAZOLE, Kemp Eliminase HG3, SULFATE ION | | 著者 | Broom, A, Rakotoharisoa, R.V, Thompson, M.C, Fraser, J.S, Chica, R.A. | | 登録日 | 2020-03-19 | | 公開日 | 2020-07-22 | | 最終更新日 | 2021-05-12 | | 実験手法 | X-RAY DIFFRACTION (1.39 Å) | | 主引用文献 | Ensemble-based enzyme design can recapitulate the effects of laboratory directed evolution in silico.
Nat Commun, 11, 2020
|
|
5RGA
 
 | | Crystal Structure of Kemp Eliminase HG3 with bound transition state analogue, 277K | | 分子名称: | 6-NITROBENZOTRIAZOLE, Kemp Eliminase HG3, SULFATE ION | | 著者 | Broom, A, Rakotoharisoa, R.V, Thompson, M.C, Fraser, J.S, Chica, R.A. | | 登録日 | 2020-03-19 | | 公開日 | 2020-07-22 | | 最終更新日 | 2021-05-12 | | 実験手法 | X-RAY DIFFRACTION (1.86 Å) | | 主引用文献 | Ensemble-based enzyme design can recapitulate the effects of laboratory directed evolution in silico.
Nat Commun, 11, 2020
|
|
5RGF
 
 | | Crystal Structure of Kemp Eliminase HG4 with bound transition state analogue, 277K | | 分子名称: | 6-NITROBENZOTRIAZOLE, Kemp Eliminase HG3, SULFATE ION | | 著者 | Broom, A, Rakotoharisoa, R.V, Thompson, M.C, Fraser, J.S, Chica, R.A. | | 登録日 | 2020-03-19 | | 公開日 | 2020-07-22 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.46 Å) | | 主引用文献 | Ensemble-based enzyme design can recapitulate the effects of laboratory directed evolution in silico.
Nat Commun, 11, 2020
|
|
5RG9
 
 | | Crystal Structure of Kemp Eliminase HG4 in unbound state, 277K | | 分子名称: | ACETATE ION, Kemp Eliminase HG4, SULFATE ION | | 著者 | Broom, A, Rakotoharisoa, R.V, Thompson, M.C, Fraser, J.S, Chica, R.A. | | 登録日 | 2020-03-19 | | 公開日 | 2020-07-22 | | 最終更新日 | 2021-05-12 | | 実験手法 | X-RAY DIFFRACTION (1.47 Å) | | 主引用文献 | Ensemble-based enzyme design can recapitulate the effects of laboratory directed evolution in silico.
Nat Commun, 11, 2020
|
|
8ATJ
 
 | | Crystal Structure of Shank2-SAM domain | | 分子名称: | CHLORIDE ION, FORMIC ACID, Isoform 4 of SH3 and multiple ankyrin repeat domains protein 2, ... | | 著者 | Bento, I, Gracia Alai, M, Kreienkamp, J.-H. | | 登録日 | 2022-08-23 | | 公開日 | 2022-11-30 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.117 Å) | | 主引用文献 | Structural deficits in key domains of Shank2 lead to alterations in postsynaptic nanoclusters and to a neurodevelopmental disorder in humans.
Mol Psychiatry, 2022
|
|
8B10
 
 | | Crystal Structure of Shank2-SAM mutant domain - L1800W | | 分子名称: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Bento, I, Gracia Alai, M, Kreienkamp, J.-H. | | 登録日 | 2022-09-08 | | 公開日 | 2022-11-30 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structural deficits in key domains of Shank2 lead to alterations in postsynaptic nanoclusters and to a neurodevelopmental disorder in humans.
Mol Psychiatry, 2022
|
|
6WL7
 
 | | Cryo-EM of Form 2 like peptide filament, 29-20-2 | | 分子名称: | peptide 29-20-2 | | 著者 | Wang, F, Gnewou, O.M, Modlin, C, Egelman, E.H, Conticello, V.P. | | 登録日 | 2020-04-18 | | 公開日 | 2020-12-02 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structural analysis of cross alpha-helical nanotubes provides insight into the designability of filamentous peptide nanomaterials.
Nat Commun, 12, 2021
|
|
7AM3
 
 | |
7AM6
 
 | |
7AM8
 
 | |
7AM4
 
 | |
7AM7
 
 | |
5K2H
 
 | |
