5W2E
 
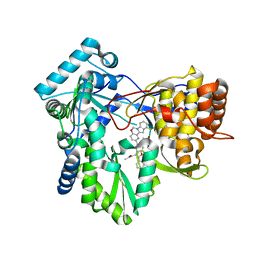 | | HCV NS5B RNA-dependent RNA polymerase in complex with non-nucleoside inhibitor MK-8876 | | 分子名称: | 2-(4-fluorophenyl)-5-(11-fluoro-6H-pyrido[2',3':5,6][1,3]oxazino[3,4-a]indol-2-yl)-N-methyl-6-[methyl(methylsulfonyl)amino]-1-benzofuran-3-carboxamide, Genome polyprotein | | 著者 | Lesburg, C.A, Ummat, A. | | 登録日 | 2017-06-06 | | 公開日 | 2017-08-16 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Development of a New Structural Class of Broadly Acting HCV Non-Nucleoside Inhibitors Leading to the Discovery of MK-8876.
ChemMedChem, 12, 2017
|
|
5FVK
 
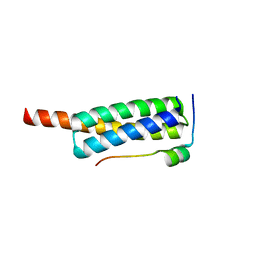 | | Crystal structure of Vps4-Vfa1 complex from S.cerevisiae at 1.66 A resolution. | | 分子名称: | VACUOLAR PROTEIN SORTING-ASSOCIATED PROTEIN 4, VPS4-ASSOCIATED PROTEIN 1 | | 著者 | Kojima, R, Obita, T, Onoue, K, Mizuguchi, M. | | 登録日 | 2016-02-09 | | 公開日 | 2016-04-27 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.658 Å) | | 主引用文献 | Structural Fine-Tuning of Mit Interacting Motif 2 (Mim2) and Allosteric Regulation of Escrt-III by Vps4 in Yeast.
J.Mol.Biol., 428, 2016
|
|
2WWA
 
 | | Cryo-EM structure of idle yeast Ssh1 complex bound to the yeast 80S ribosome | | 分子名称: | 25S RRNA, 60S RIBOSOMAL PROTEIN L17-A, 60S RIBOSOMAL PROTEIN L19, ... | | 著者 | Becker, T, Mandon, E, Bhushan, S, Jarasch, A, Armache, J.P, Funes, S, Jossinet, F, Gumbart, J, Mielke, T, Berninghausen, O, Schulten, K, Westhof, E, Gilmore, R, Beckmann, R. | | 登録日 | 2009-10-22 | | 公開日 | 2009-12-08 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (8.9 Å) | | 主引用文献 | Structure of Monomeric Yeast and Mammalian Sec61 Complexes Interacting with the Translating Ribosome.
Science, 326, 2009
|
|
5FVL
 
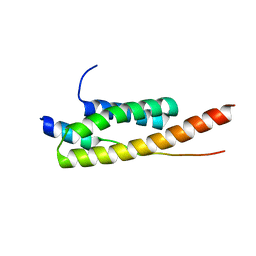 | | Crystal structure of Vps4-Vps20 complex from S.cerevisiae | | 分子名称: | VACUOLAR PROTEIN SORTING-ASSOCIATED PROTEIN 20, VACUOLAR PROTEIN SORTING-ASSOCIATED PROTEIN 4 | | 著者 | Kojima, R, Obita, T, Onoue, K, Mizuguchi, M. | | 登録日 | 2016-02-09 | | 公開日 | 2016-04-27 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.973 Å) | | 主引用文献 | Structural Fine-Tuning of Mit Interacting Motif 2 (Mim2) and Allosteric Regulation of Escrt-III by Vps4 in Yeast.
J.Mol.Biol., 428, 2016
|
|
2WW9
 
 | | Cryo-EM structure of the active yeast Ssh1 complex bound to the yeast 80S ribosome | | 分子名称: | 25S RRNA, 60S RIBOSOMAL PROTEIN L17-A, 60S RIBOSOMAL PROTEIN L19, ... | | 著者 | Becker, T, Mandon, E, Bhushan, S, Jarasch, A, Armache, J.P, Funes, S, Jossinet, F, Gumbart, J, Mielke, T, Berninghausen, O, Schulten, K, Westhof, E, Gilmore, R, Beckmann, R. | | 登録日 | 2009-10-22 | | 公開日 | 2009-12-08 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (8.6 Å) | | 主引用文献 | Structure of Monomeric Yeast and Mammalian Sec61 Complexes Interacting with the Translating Ribosome.
Science, 326, 2009
|
|
2WWB
 
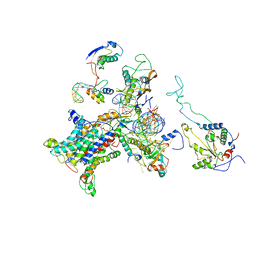 | | CRYO-EM STRUCTURE OF THE MAMMALIAN SEC61 COMPLEX BOUND TO THE ACTIVELY TRANSLATING WHEAT GERM 80S RIBOSOME | | 分子名称: | 25S RRNA, 5.8S RRNA, 60S RIBOSOMAL PROTEIN L17-A, ... | | 著者 | Becker, T, Mandon, E, Bhushan, S, Jarasch, A, Armache, J.P, Funes, S, Jossinet, F, Gumbart, J, Mielke, T, Berninghausen, O, Schulten, K, Westhof, E, Gilmore, R, Beckmann, R. | | 登録日 | 2009-10-22 | | 公開日 | 2009-12-08 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (6.48 Å) | | 主引用文献 | Structure of Monomeric Yeast and Mammalian Sec61 Complexes Interacting with the Translating Ribosome.
Science, 326, 2009
|
|
5Z0U
 
 | |
5Z0T
 
 | |
2MVT
 
 | | Solution structure of scoloptoxin SSD609 from Scolopendra mutilans | | 分子名称: | Scoloptoxin SSD609 | | 著者 | Wu, F, Sun, P, Wang, C, He, Y, Zhang, L, Tian, C. | | 登録日 | 2014-10-14 | | 公開日 | 2015-09-23 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | A distinct three-helix centipede toxin SSD609 inhibits Iks channels by interacting with the KCNE1 auxiliary subunit.
Sci Rep, 5, 2015
|
|
8F7H
 
 | | The condensation domain of surfactin A synthetase C variant 18b in space group P212121 | | 分子名称: | GLYCEROL, Surfactin synthetase | | 著者 | Frota, N.F, Pistofidis, A, Folger, I.B, Hilvert, D, Schmeing, M. | | 登録日 | 2022-11-18 | | 公開日 | 2023-11-22 | | 最終更新日 | 2024-06-12 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | High-throughput reprogramming of an NRPS condensation domain.
Nat.Chem.Biol., 20, 2024
|
|
8F7G
 
 | | The condensation domain of surfactin A synthetase C in space group P212121 | | 分子名称: | GLYCEROL, Surfactin synthetase | | 著者 | Frota, N.F, Pistofidis, A, Folger, I.B, Hilvert, D, Schmeing, T.M. | | 登録日 | 2022-11-18 | | 公開日 | 2023-11-22 | | 最終更新日 | 2024-06-12 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | High-throughput reprogramming of an NRPS condensation domain.
Nat.Chem.Biol., 20, 2024
|
|
8F7I
 
 | | The condensation domain of surfactin A synthetase C variant 18b in space group P43212 | | 分子名称: | GLYCEROL, Surfactin synthetase | | 著者 | Frota, N.F, Pistofidis, A, Folger, I.B, Hilvert, D, Schmeing, M. | | 登録日 | 2022-11-18 | | 公開日 | 2023-11-22 | | 最終更新日 | 2024-06-12 | | 実験手法 | X-RAY DIFFRACTION (1.58 Å) | | 主引用文献 | High-throughput reprogramming of an NRPS condensation domain.
Nat.Chem.Biol., 20, 2024
|
|
8F7F
 
 | | The condensation domain of surfactin A synthetase C in space group P43212 | | 分子名称: | GLYCEROL, Surfactin synthetase | | 著者 | Frota, N, Pistofidis, A, Folger, I.B, Hilvert, D, Schmeing, T.M. | | 登録日 | 2022-11-18 | | 公開日 | 2023-11-22 | | 最終更新日 | 2024-06-12 | | 実験手法 | X-RAY DIFFRACTION (1.62 Å) | | 主引用文献 | High-throughput reprogramming of an NRPS condensation domain.
Nat.Chem.Biol., 20, 2024
|
|
6B9H
 
 | |
5FGM
 
 | | Streptomyces coelicolor SigR region 4 | | 分子名称: | ECF RNA polymerase sigma factor SigR | | 著者 | Park, S.Y. | | 登録日 | 2015-12-21 | | 公開日 | 2016-03-02 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | In Streptomyces coelicolor SigR, methionine at the -35 element interacting region 4 confers the -31'-adenine base selectivity
Biochem.Biophys.Res.Commun., 470, 2016
|
|
8PR1
 
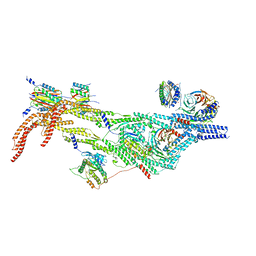 | | Cytoplasmic dynein-B heavy chain bound to IC-LC tower | | 分子名称: | Cytoplasmic dynein 1 heavy chain 1, Cytoplasmic dynein 1 intermediate chain 2, Cytoplasmic dynein 1 light intermediate chain 2, ... | | 著者 | Singh, K, Lau, C.K, Manigrasso, G, Gassmann, R, Carter, A.P. | | 登録日 | 2023-07-12 | | 公開日 | 2024-03-27 | | 最終更新日 | 2024-04-10 | | 実験手法 | ELECTRON MICROSCOPY (8.2 Å) | | 主引用文献 | Molecular mechanism of dynein-dynactin complex assembly by LIS1.
Science, 383, 2024
|
|
8PQV
 
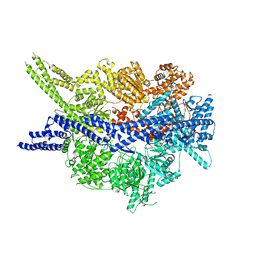 | | Cytoplasmic dynein-1 motor domain in post-powerstroke state | | 分子名称: | Cytoplasmic dynein 1 heavy chain 1 | | 著者 | Singh, K, Lau, C.K, Manigrasso, G, Gassmann, R, Carter, A.P. | | 登録日 | 2023-07-12 | | 公開日 | 2024-03-27 | | 最終更新日 | 2024-04-10 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Molecular mechanism of dynein-dynactin complex assembly by LIS1.
Science, 383, 2024
|
|
5L44
 
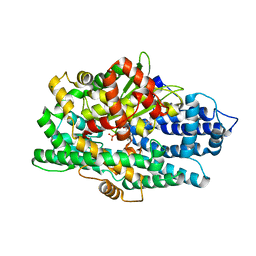 | | Structure of K-26-DCP in complex with the K-26 tripeptide | | 分子名称: | K-26 dipeptidyl carboxypeptidase, MAGNESIUM ION, N-ACETYL-L-ILE-L-TYR-(R)-1-AMINO-2-(4-HYDROXYPHENYL)ETHYLPHOSPHONIC ACID, ... | | 著者 | Masuyer, G, Acharya, K.R, Kramer, G.J, Bachmann, B.O. | | 登録日 | 2016-05-24 | | 公開日 | 2016-10-26 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Crystal structure of a peptidyl-dipeptidase K-26-DCP from Actinomycete in complex with its natural inhibitor.
FEBS J., 283, 2016
|
|
5DTM
 
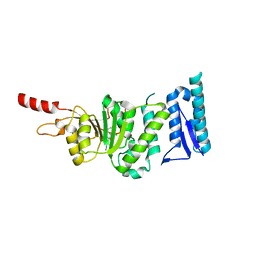 | | Crystal structure of Dot1L in complex with inhibitor CPD1 [4-(2,6-dichlorobenzoyl)-N-methyl-1H-pyrrole-2-carboxamide] | | 分子名称: | 4-(2,6-dichlorobenzoyl)-N-methyl-1H-pyrrole-2-carboxamide, Histone-lysine N-methyltransferase, H3 lysine-79 specific | | 著者 | Scheufler, C, Be, C, Moebitz, H, Stauffer, F. | | 登録日 | 2015-09-18 | | 公開日 | 2016-06-15 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Optimization of a Fragment-Based Screening Hit toward Potent DOT1L Inhibitors Interacting in an Induced Binding Pocket.
Acs Med.Chem.Lett., 7, 2016
|
|
5EHK
 
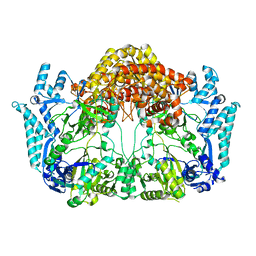 | |
3NMP
 
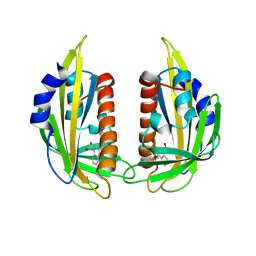 | | Crystal structure of the abscisic receptor PYL2 mutant A93F in complex with pyrabactin | | 分子名称: | 4-bromo-N-(pyridin-2-ylmethyl)naphthalene-1-sulfonamide, Abscisic acid receptor PYL2 | | 著者 | Zhou, X.E, Melcher, K, Ng, L.-M, Soon, F.-F, Xu, Y, Suino-Powell, K.M, Kovach, A, Li, J, Yong, E.-L, Xu, H.E. | | 登録日 | 2010-06-22 | | 公開日 | 2010-08-25 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Identification and mechanism of ABA receptor antagonism.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3NMT
 
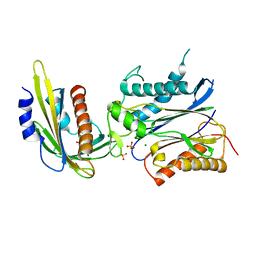 | | Crystal structure of pyrabactin bound abscisic acid receptor PYL2 mutant A93F in complex with type 2C protein phosphatase HAB1 | | 分子名称: | 4-bromo-N-(pyridin-2-ylmethyl)naphthalene-1-sulfonamide, Abscisic acid receptor PYL2, MAGNESIUM ION, ... | | 著者 | Zhou, X.E, Melcher, K, Ng, L.-M, Soon, F.-F, Xu, Y, Suino-Powell, K.M, Kovach, A, Li, J, Yong, E.-L, Xu, H.E. | | 登録日 | 2010-06-22 | | 公開日 | 2010-08-25 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.56 Å) | | 主引用文献 | Identification and mechanism of ABA receptor antagonism.
Nat.Struct.Mol.Biol., 17, 2010
|
|
5DTQ
 
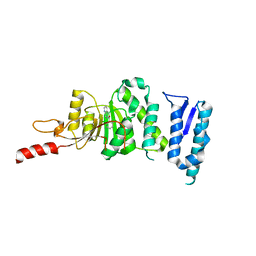 | | Crystal structure of Dot1L in complex with inhibitor CPD3 [(2,6-dichlorophenyl)(quinolin-6-yl)methanone] | | 分子名称: | (2,6-dichlorophenyl)(quinolin-6-yl)methanone, Histone-lysine N-methyltransferase, H3 lysine-79 specific | | 著者 | Scheufler, C, Be, C, Moebitz, H, Stauffer, F. | | 登録日 | 2015-09-18 | | 公開日 | 2016-06-15 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.61 Å) | | 主引用文献 | Optimization of a Fragment-Based Screening Hit toward Potent DOT1L Inhibitors Interacting in an Induced Binding Pocket.
Acs Med.Chem.Lett., 7, 2016
|
|
3NMV
 
 | | Crystal structure of pyrabactin-bound abscisic acid receptor PYL2 mutant A93F in complex with type 2C protein phosphatase ABI2 | | 分子名称: | 4-bromo-N-(pyridin-2-ylmethyl)naphthalene-1-sulfonamide, Abscisic acid receptor PYL2, MAGNESIUM ION, ... | | 著者 | Zhou, X.E, Melcher, K, Ng, L.-M, Soon, F.-F, Xu, Y, Suino-Powell, K.M, Kovach, A, Li, J, Yong, E.-L, Xu, H.E. | | 登録日 | 2010-06-22 | | 公開日 | 2010-08-25 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Identification and mechanism of ABA receptor antagonism.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3NMN
 
 | | Crystal structure of pyrabactin-bound abscisic acid receptor PYL1 in complex with type 2C protein phosphatase ABI1 | | 分子名称: | 4-bromo-N-(pyridin-2-ylmethyl)naphthalene-1-sulfonamide, Abscisic acid receptor PYL1, MAGNESIUM ION, ... | | 著者 | Zhou, X.E, Melcher, K, Ng, L.-M, Soon, F.-F, Xu, Y, Suino-Powell, K.M, Kovach, A, Li, J, Yong, E.-L, Xu, H.E. | | 登録日 | 2010-06-22 | | 公開日 | 2010-08-25 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Identification and mechanism of ABA receptor antagonism.
Nat.Struct.Mol.Biol., 17, 2010
|
|
