7S7Y
 
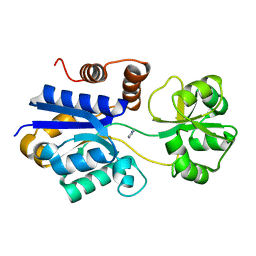 | | Crystal structure of iCytSnFR Cytisine Sensor precursor binding protein | | 分子名称: | IMIDAZOLE, iNicSnFR 4.0 Fluorescent Nicotine Sensor precursor binding protein | | 著者 | Fan, C, Nichols, N.L, Luebbert, L, Looger, L.L, Lester, H.A, Rees, D.C. | | 登録日 | 2021-09-17 | | 公開日 | 2021-10-13 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Structure, Function, and Application of Bacterial ABC Transporters
Ph.D.Thesis,California Institute of Technology, 2020
|
|
7SAH
 
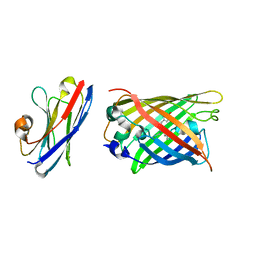 | |
7SAI
 
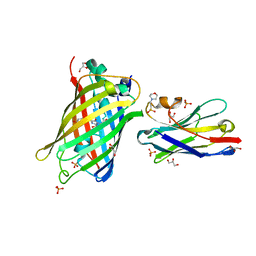 | |
7SAJ
 
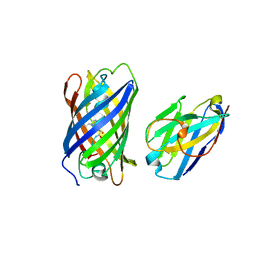 | |
7SAK
 
 | |
7SAL
 
 | |
7SF9
 
 | |
7SFA
 
 | |
7SQC
 
 | | Ciliary C1 central pair apparatus isolated from Chlamydomonas reinhardtii | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, CPC1, Calmodulin, ... | | 著者 | Gui, M, Wang, X, Dutcher, S.K, Brown, A, Zhang, R. | | 登録日 | 2021-11-05 | | 公開日 | 2022-04-13 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Ciliary central apparatus structure reveals mechanisms of microtubule patterning.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7SQY
 
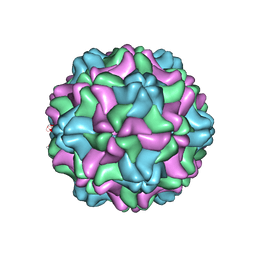 | | CSDaV GFP mutant | | 分子名称: | Citrus Sudden Death-associated Virus Capsid Protein,Green fluorescent protein,Citrus Sudden Death-associated Virus Capsid Protein | | 著者 | Guo, F, Matsumura, E.E, Falk, B.W. | | 登録日 | 2021-11-07 | | 公開日 | 2022-05-25 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Citrus sudden death-associated virus as a new expression vector for rapid in planta production of heterologous proteins, chimeric virions, and virus-like particles.
Biotechnol Rep., 35, 2022
|
|
7SQZ
 
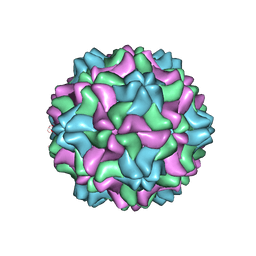 | | CSDaV wild-type | | 分子名称: | Citrus Sudden Death-associated Virus Capsid Protein | | 著者 | Guo, F, Matsumura, E.E, Falk, B.W. | | 登録日 | 2021-11-07 | | 公開日 | 2022-05-25 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Citrus sudden death-associated virus as a new expression vector for rapid in planta production of heterologous proteins, chimeric virions, and virus-like particles.
Biotechnol Rep., 35, 2022
|
|
7SSV
 
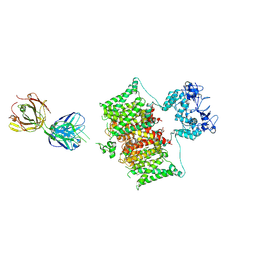 | | Structure of human Kv1.3 with Fab-ShK fusion | | 分子名称: | Fab-ShK fusion, heavy chain, light chain, ... | | 著者 | Meyerson, J.R, Selvakumar, P, Smider, V, Huang, R. | | 登録日 | 2021-11-11 | | 公開日 | 2022-06-29 | | 最終更新日 | 2022-07-20 | | 実験手法 | ELECTRON MICROSCOPY (3.39 Å) | | 主引用文献 | Structures of the T cell potassium channel Kv1.3 with immunoglobulin modulators.
Nat Commun, 13, 2022
|
|
7SSX
 
 | | Structure of human Kv1.3 | | 分子名称: | POTASSIUM ION, Potassium voltage-gated channel subfamily A member 3, Green fluorescent protein fusion | | 著者 | Meyerson, J.R, Selvakumar, P. | | 登録日 | 2021-11-11 | | 公開日 | 2022-06-29 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (2.89 Å) | | 主引用文献 | Structures of the T cell potassium channel Kv1.3 with immunoglobulin modulators.
Nat Commun, 13, 2022
|
|
7SSY
 
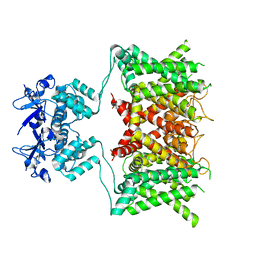 | |
7SSZ
 
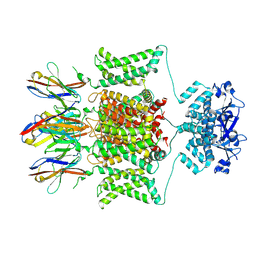 | |
7SUK
 
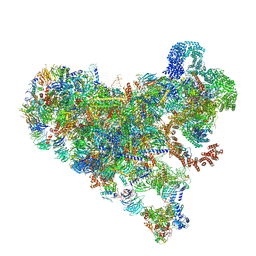 | | Structure of Bfr2-Lcp5 Complex Observed in the Small Subunit Processome Isolated from R2TP-depleted Yeast Cells | | 分子名称: | 18S pre-rRNA, 40S ribosomal protein S11-A, 40S ribosomal protein S13, ... | | 著者 | Rai, J, Zhao, Y, Li, H. | | 登録日 | 2021-11-17 | | 公開日 | 2022-07-06 | | 最終更新日 | 2023-08-16 | | 実験手法 | ELECTRON MICROSCOPY (3.99 Å) | | 主引用文献 | Artificial intelligence-assisted cryoEM structure of Bfr2-Lcp5 complex observed in the yeast small subunit processome.
Commun Biol, 5, 2022
|
|
7SX3
 
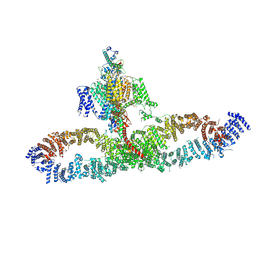 | | Human NALCN-FAM155A-UNC79-UNC80 channelosome with CaM bound, conformation 1/2 | | 分子名称: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Kschonsak, M, Chua, H.C, Weidling, C, Chakouri, N, Noland, C.L, Schott, K, Chang, T, Tam, C, Patel, N, Arthur, C.P, Leitner, A, Ben-Johny, M, Ciferri, C, Pless, S.A, Payandeh, J. | | 登録日 | 2021-11-22 | | 公開日 | 2021-12-29 | | 最終更新日 | 2022-03-16 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structural architecture of the human NALCN channelosome.
Nature, 603, 2022
|
|
7SX4
 
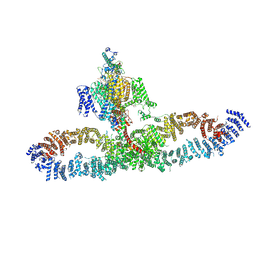 | | Human NALCN-FAM155A-UNC79-UNC80 channelosome with CaM bound, conformation 2/2 | | 分子名称: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Kschonsak, M, Chua, H.C, Weidling, C, Chakouri, N, Noland, C.L, Schott, K, Chang, T, Tam, C, Patel, N, Arthur, C.P, Leitner, A, Ben-Johny, M, Ciferri, C, Pless, S.A, Payandeh, J. | | 登録日 | 2021-11-22 | | 公開日 | 2021-12-29 | | 最終更新日 | 2022-03-16 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structural architecture of the human NALCN channelosome.
Nature, 603, 2022
|
|
7SYG
 
 | | Structure of the HCV IRES binding to the 40S ribosomal subunit, closed conformation. Structure 1(delta dII) | | 分子名称: | 18S rRNA, 40S ribosomal protein S2, 40S ribosomal protein S24, ... | | 著者 | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | 登録日 | 2021-11-25 | | 公開日 | 2022-07-13 | | 最終更新日 | 2024-02-28 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | Molecular architecture of 40S initiation complexes on the Hepatitis C virus IRES: from ribosomal attachment to eIF5B-mediated reorientation of initiator tRNA
To Be Published
|
|
7SYH
 
 | | Structure of the HCV IRES binding to the 40S ribosomal subunit, closed conformation. Structure 2(delta dII) | | 分子名称: | 18S rRNA, 40S ribosomal protein S21, 40S ribosomal protein S24, ... | | 著者 | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | 登録日 | 2021-11-25 | | 公開日 | 2022-07-13 | | 実験手法 | ELECTRON MICROSCOPY (4.6 Å) | | 主引用文献 | Molecular architecture of 40S initiation complexes on the Hepatitis C virus IRES: from ribosomal attachment to eIF5B-mediated reorientation of initiator tRNA
To Be Published
|
|
7SYI
 
 | | Structure of the HCV IRES binding to the 40S ribosomal subunit, closed conformation. Structure 3(delta dII) | | 分子名称: | 18S rRNA, 40S ribosomal protein S21, 40S ribosomal protein S24, ... | | 著者 | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | 登録日 | 2021-11-25 | | 公開日 | 2022-07-13 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES.
Embo J., 41, 2022
|
|
7SYJ
 
 | | Structure of the HCV IRES binding to the 40S ribosomal subunit, closed conformation. Structure 4(delta dII) | | 分子名称: | 18S rRNA, 40S ribosomal protein S21, 40S ribosomal protein S24, ... | | 著者 | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | 登録日 | 2021-11-25 | | 公開日 | 2022-07-13 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (4.8 Å) | | 主引用文献 | Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES.
Embo J., 41, 2022
|
|
7SYK
 
 | | Structure of the HCV IRES binding to the 40S ribosomal subunit, closed conformation. Structure 5(delta dII) | | 分子名称: | 18S rRNA, 40S ribosomal protein S21, 40S ribosomal protein S24, ... | | 著者 | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | 登録日 | 2021-11-25 | | 公開日 | 2022-07-13 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES.
Embo J., 41, 2022
|
|
7SYL
 
 | | Structure of the HCV IRES bound to the 40S ribosomal subunit, closed conformation. Structure 6(delta dII) | | 分子名称: | 18S rRNA, 40S ribosomal protein S21, 40S ribosomal protein S24, ... | | 著者 | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | 登録日 | 2021-11-25 | | 公開日 | 2022-07-13 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES.
Embo J., 41, 2022
|
|
7SYM
 
 | | Structure of the HCV IRES bound to the 40S ribosomal subunit, head opening. Structure 7(delta dII) | | 分子名称: | 18S rRNA, 40S ribosomal protein S2, 40S ribosomal protein S21, ... | | 著者 | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | 登録日 | 2021-11-25 | | 公開日 | 2022-07-13 | | 最終更新日 | 2024-02-28 | | 実験手法 | ELECTRON MICROSCOPY (4.8 Å) | | 主引用文献 | Comprehensive structural overview of the HCV IRES-mediated translation initiation pathway
To Be Published
|
|
