7MDT
 
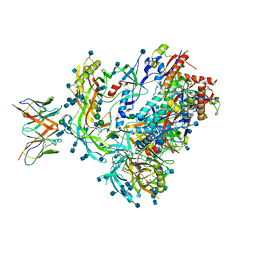 | | BG505 SOSIP.v5.2 in complex with the monoclonal antibody Rh4O9.8 (as Fab fragment) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Rh4O9.8 monoclonal antibody Heavy Chain, ... | | 著者 | Antanasijevic, A, Ozorowski, G, Nogal, B, Ward, A.B. | | 登録日 | 2021-04-06 | | 公開日 | 2022-01-26 | | 最終更新日 | 2022-02-02 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | From structure to sequence: Antibody discovery using cryoEM.
Sci Adv, 8, 2022
|
|
6D69
 
 | |
2Y64
 
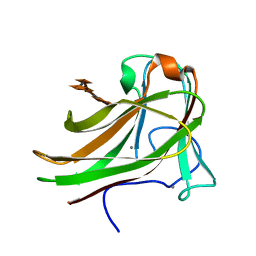 | | Xylopentaose binding mutated (X-2 L110F) CBM4-2 Carbohydrate Binding Module from a Thermostable Rhodothermus marinus Xylanase | | 分子名称: | CALCIUM ION, XYLANASE, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | 著者 | von Schantz, L, Hakansson, M, Logan, D.T, Walse, B, Osterlin, J, Nordberg-Karlsson, E, Ohlin, M. | | 登録日 | 2011-01-19 | | 公開日 | 2012-03-07 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structural basis for carbohydrate-binding specificity--a comparative assessment of two engineered carbohydrate-binding modules.
Glycobiology, 22, 2012
|
|
2YV9
 
 | |
7OS7
 
 | | Circular permutant of ribosomal protein S6, swap helix 2, L75A, A92K mutant | | 分子名称: | 30S ribosomal protein S6,30S ribosomal protein S6 | | 著者 | Wang, H, Logan, D.T, Oliveberg, M. | | 登録日 | 2021-06-08 | | 公開日 | 2022-06-22 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Circular permutant of ribosomal protein S6, swap helix 2, L75A, A92K mutant
To Be Published
|
|
5WFE
 
 | | Cas1-Cas2-IHF-DNA holo-complex | | 分子名称: | CRISPR-associated endonuclease Cas1, CRISPR-associated endoribonuclease Cas2, DNA (28-MER), ... | | 著者 | Wright, A.V, Liu, J.J, Nogales, E, Doudna, J.A. | | 登録日 | 2017-07-11 | | 公開日 | 2017-08-02 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (3.64 Å) | | 主引用文献 | Structures of the CRISPR genome integration complex.
Science, 357, 2017
|
|
2YOA
 
 | | Synaptotagmin-1 C2B domain with phosphoserine | | 分子名称: | CALCIUM ION, PHOSPHOSERINE, SYNAPTOTAGMIN-1, ... | | 著者 | Honigmann, A, van den Bogaart, G, Iraheta, E, Risselada, H.J, Milovanovic, D, Mueller, V, Muellar, S, Diederichsen, U, Fasshauer, D, Grubmuller, H, Hell, S.W, Eggeling, C, Kuhnel, K, Jahn, R. | | 登録日 | 2012-10-22 | | 公開日 | 2013-03-20 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Phosphatidylinositol 4,5-Bisphosphate Clusters Act as Molecular Beacons for Vesicle Recruitment
Nat.Struct.Mol.Biol., 20, 2013
|
|
2Y6G
 
 | | Cellopentaose binding mutated (X-2 L110F) CBM4-2 Carbohydrate Binding Module from a Thermostable Rhodothermus marinus Xylanase | | 分子名称: | CALCIUM ION, XYLANASE, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | 著者 | von Schantz, L, Hakansson, M, Logan, D.T, Walse, B, Osterlin, J, Nordberg-Karlsson, E, Ohlin, M. | | 登録日 | 2011-01-21 | | 公開日 | 2012-03-07 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Structural basis for carbohydrate-binding specificity--a comparative assessment of two engineered carbohydrate-binding modules.
Glycobiology, 22, 2012
|
|
1RW9
 
 | | Crystal structure of the Arthrobacter aurescens chondroitin AC lyase | | 分子名称: | PHOSPHATE ION, SODIUM ION, chondroitin AC lyase | | 著者 | Lunin, V.V, Li, Y, Linhardt, R.J, Miyazono, H, Kyogashima, M, Kaneko, T, Bell, A.W, Cygler, M. | | 登録日 | 2003-12-16 | | 公開日 | 2004-04-13 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | High-resolution crystal structure of Arthrobacter aurescens chondroitin AC lyase: an enzyme-substrate complex defines the catalytic mechanism
J.Mol.Biol., 337, 2004
|
|
2YV0
 
 | | Structural and Thermodynamic Analyses of E. coli ribonuclease HI Variant with Quintuple Thermostabilizing Mutations | | 分子名称: | Ribonuclease HI | | 著者 | Haruki, M, Motegi, T, Tadokoro, T, Koga, Y, Takano, K, Kanaya, S. | | 登録日 | 2007-04-06 | | 公開日 | 2008-03-18 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structural and thermodynamic analyses of Escherichia coli RNase HI variant with quintuple thermostabilizing mutations.
Febs J., 274, 2007
|
|
4UFQ
 
 | | Structure of a novel Hyaluronidase (Hyal_Sk) from Streptomyces koganeiensis. | | 分子名称: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | 著者 | Gavira, J.A, Messina, L, Pernagallo, S, Unciti-Broceta, J.D, Conejero-Muriel, M, Diaz-Mochon, J.J, Vaccaro, S, Caruso, S, Musumeci, L, Bisicchia, S, Di Pasquale, R. | | 登録日 | 2015-03-18 | | 公開日 | 2016-04-13 | | 最終更新日 | 2017-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Identification and Characterization of a Bacterial Hyaluronidase and its Production in Recombinant Form.
FEBS Lett., 590, 2016
|
|
1RHH
 
 | | Crystal Structure of the Broadly HIV-1 Neutralizing Fab X5 at 1.90 Angstrom Resolution | | 分子名称: | Fab X5, heavy chain, light chain | | 著者 | Darbha, R, Phogat, S, Labrijn, A.F, Shu, Y, Gu, Y, Andrykovitch, M, Zhang, M.Y, Pantophlet, R, Martin, L, Vita, C, Burton, D.R, Dimitrov, D.S, Ji, X. | | 登録日 | 2003-11-14 | | 公開日 | 2004-02-24 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal Structure of the Broadly Cross-Reactive HIV-1-Neutralizing Fab X5 and Fine Mapping of Its Epitope
Biochemistry, 43, 2004
|
|
7PQE
 
 | |
7PPO
 
 | |
2Y6L
 
 | | Xylopentaose binding X-2 engineered mutated CBM4-2 Carbohydrate Binding Module from a Thermostable Rhodothermus marinus Xylanase | | 分子名称: | CALCIUM ION, XYLANASE, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | 著者 | von Schantz, L, Hakansson, M, Logan, D.T, Walse, B, Osterlin, J, Nordberg-Karlsson, E, Ohlin, M. | | 登録日 | 2011-01-24 | | 公開日 | 2012-03-07 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.28 Å) | | 主引用文献 | Structural basis for carbohydrate-binding specificity--a comparative assessment of two engineered carbohydrate-binding modules.
Glycobiology, 22, 2012
|
|
1RWF
 
 | | Crystal structure of Arthrobacter aurescens chondroitin AC lyase in complex with chondroitin tetrasaccharide | | 分子名称: | 2,6-anhydro-3-deoxy-L-threo-hex-2-enonic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-2,6-anhydro-3-deoxy-L-xylo-hexonic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose, PHOSPHATE ION, SODIUM ION, ... | | 著者 | Lunin, V.V, Li, Y, Miyazono, H, Kyogashima, M, Bell, A.W, Cygler, M. | | 登録日 | 2003-12-16 | | 公開日 | 2004-04-13 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | High-resolution crystal structure of Arthrobacter aurescens chondroitin AC lyase: an enzyme-substrate complex defines the catalytic mechanism
J.Mol.Biol., 337, 2004
|
|
1RWG
 
 | | Crystal structure of Arthrobacter aurescens chondroitin AC lyase in complex with chondroitin tetrasaccharide | | 分子名称: | 2,6-anhydro-3-deoxy-L-threo-hex-2-enonic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-2,6-anhydro-3-deoxy-L-xylo-hexonic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose, PHOSPHATE ION, SODIUM ION, ... | | 著者 | Lunin, V.V, Li, Y, Miyazono, H, Kyogashima, M, Bell, A.W, Cygler, M. | | 登録日 | 2003-12-16 | | 公開日 | 2004-04-13 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | High-resolution crystal structure of Arthrobacter aurescens chondroitin AC lyase: an enzyme-substrate complex defines the catalytic mechanism
J.Mol.Biol., 337, 2004
|
|
6QFF
 
 | | Crystal Structure of Human Kallikrein 6 in complex with GSK144 | | 分子名称: | 4-[(5-phenyl-1~{H}-imidazol-2-yl)methylamino]-2-(pyridin-3-ylmethoxy)benzenecarboximidamide, GLYCEROL, Kallikrein-6 | | 著者 | Thorpe, J.H. | | 登録日 | 2019-01-10 | | 公開日 | 2019-05-08 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.64 Å) | | 主引用文献 | Evaluation of a crystallographic surrogate for kallikrein 5 in the discovery of novel inhibitors for Netherton syndrome.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
7BQE
 
 | | Solution NMR structure of NF3; de novo designed protein with a novel fold | | 分子名称: | NF3 | | 著者 | Kobayashi, N, Sugiki, T, Fujiwara, T, Minami, S, Koga, R, Chikenji, G, Koga, N. | | 登録日 | 2020-03-24 | | 公開日 | 2021-03-24 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Exploration of novel alpha-beta-protein folds through de novo design
Nat.Struct.Mol.Biol., 2023
|
|
7BQQ
 
 | | Solution NMR structure of fold-Z Gogy; de novo designed protein with an asymmetric all-alpha topology | | 分子名称: | Gogy | | 著者 | Kobayashi, N, Sugiki, T, Fujiwara, T, Sakuma, K, Kosugi, T, Koga, R, Koga, N. | | 登録日 | 2020-03-25 | | 公開日 | 2021-04-07 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Design of complicated all-alpha protein structures
Nat.Struct.Mol.Biol., 2024
|
|
7BQM
 
 | | Solution NMR structure of fold-0 Chantal; de novo designed protein with an asymmetric all-alpha topology | | 分子名称: | Chantal | | 著者 | Kobayashi, N, Sugiki, T, Fujiwara, T, Sakuma, K, Kosugi, T, Koga, R, Koga, N. | | 登録日 | 2020-03-25 | | 公開日 | 2021-04-07 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Design of complicated all-alpha protein structures
Nat.Struct.Mol.Biol., 2024
|
|
7BQB
 
 | | Solution NMR structure of NF6; de novo designed protein with a novel fold | | 分子名称: | NF6 | | 著者 | Kobayashi, N, Nagashima, T, Minami, S, Koga, R, Chikenji, G, Koga, N. | | 登録日 | 2020-03-24 | | 公開日 | 2021-03-24 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Exploration of novel alpha-beta-protein folds through de novo design
Nat.Struct.Mol.Biol., 2023
|
|
7BQC
 
 | | Solution NMR structure of NF4; de novo designed protein with a novel fold | | 分子名称: | NF4 | | 著者 | Kobayashi, N, Nagashima, T, Minami, S, Koga, R, Chikenji, T, Koga, N. | | 登録日 | 2020-03-24 | | 公開日 | 2021-03-24 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Exploration of novel alpha-beta-protein folds through de novo design
Nat.Struct.Mol.Biol., 2023
|
|
7BQN
 
 | | Solution NMR structure of fold-C Rei; de novo designed protein with an asymmetric all-alpha topology | | 分子名称: | Rei | | 著者 | Kobayashi, N, Sugiki, T, Fujiwara, T, Sakuma, K, Kosugi, T, Koga, R, Koga, N. | | 登録日 | 2020-03-25 | | 公開日 | 2021-04-07 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Design of complicated all-alpha protein structures
Nat.Struct.Mol.Biol., 2024
|
|
1FGX
 
 | | CRYSTAL STRUCTURE OF THE BOVINE BETA 1,4 GALACTOSYLTRANSFERASE (B4GALT1) CATALYTIC DOMAIN COMPLEXED WITH UMP | | 分子名称: | BETA 1,4 GALACTOSYLTRANSFERASE, URIDINE-5'-MONOPHOSPHATE | | 著者 | Gastinel, L.N, Cambillau, C, Bourne, Y. | | 登録日 | 2000-07-29 | | 公開日 | 2000-08-16 | | 最終更新日 | 2021-07-21 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structures of the bovine beta4galactosyltransferase catalytic domain and its complex with uridine diphosphogalactose.
EMBO J., 18, 1999
|
|
