3ME4
 
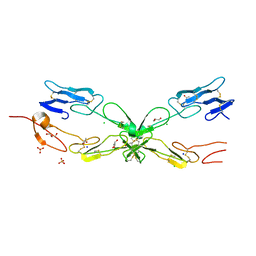 | | Crystal structure of mouse RANK | | 分子名称: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | 著者 | Walter, S.W, Liu, C, Zhu, X, Wu, Y, Owens, R.J, Stuart, D.I, Gao, B, Ren, J. | | 登録日 | 2010-03-31 | | 公開日 | 2010-06-02 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | Structural and Functional Insights of RANKL-RANK Interaction and Signaling.
J.Immunol., 2010
|
|
3MJQ
 
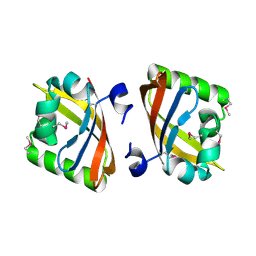 | | Crystal structure of the PAS domain of Q24QT8_DESHY protein from Desulfitobacterium hafniense. Northeast Structural Genomics Consortium Target DhR85c. | | 分子名称: | uncharacterized protein | | 著者 | Vorobiev, S, Neely, H, Seetharaman, J, Wang, D, Ciccosanti, C, Mao, L, Xiao, R, Acton, T.B, Everett, J.K, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2010-04-13 | | 公開日 | 2010-04-28 | | 最終更新日 | 2021-10-06 | | 実験手法 | X-RAY DIFFRACTION (2.601 Å) | | 主引用文献 | Crystal structure of the PAS domain of Q24QT8_DESHY protein from Desulfitobacterium hafniense.
To be Published
|
|
1R3D
 
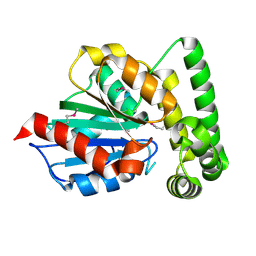 | |
3MGK
 
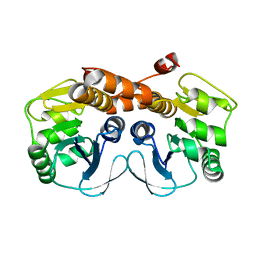 | | CRYSTAL STRUCTURE OF PROBABLE PROTEASE/AMIDASE FROM Clostridium acetobutylicum ATCC 824 | | 分子名称: | Intracellular protease/amidase related enzyme (ThiJ family) | | 著者 | Patskovsky, Y, Toro, R, Freeman, J, Iizuka, M, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2010-04-06 | | 公開日 | 2010-04-21 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | CRYSTAL STRUCTURE OF PROBABLE PROTEASE/AMIDASE FROM Clostridium acetobutylicum
To be Published
|
|
3MNH
 
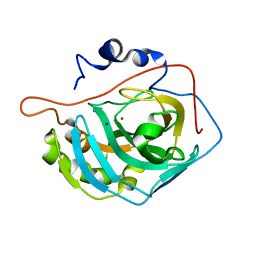 | | Human Carbonic Anhydrase II Mutant K170A | | 分子名称: | Carbonic anhydrase 2, SODIUM ION, ZINC ION | | 著者 | Domsic, J.F, McKenna, R. | | 登録日 | 2010-04-21 | | 公開日 | 2010-07-14 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Structural and kinetic study of the extended active site for proton transfer in human carbonic anhydrase II.
Biochemistry, 49, 2010
|
|
1R9G
 
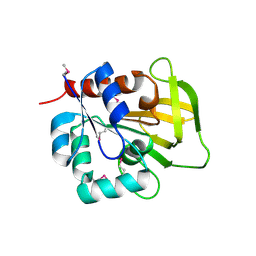 | | Three-dimensional Structure of YaaE from Bacillus subtilis | | 分子名称: | Hypothetical protein yaaE | | 著者 | Bauer, J.A, Bennett, E.M, Begley, T.P, Ealick, S.E. | | 登録日 | 2003-10-29 | | 公開日 | 2004-01-27 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Three-dimensional Structure of YaaE from Bacillus subtilis, a Glutaminase Implicated in Pyridoxal-5'-phosphate Biosynthesis.
J.Biol.Chem., 279, 2004
|
|
1R5G
 
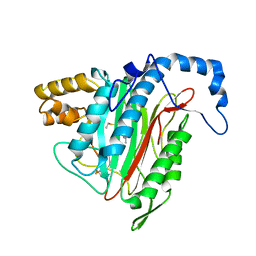 | | Crystal Structure of MetAP2 complexed with A311263 | | 分子名称: | (2S,3R)-3-AMINO-2-HYDROXY-5-(ETHYLSULFANYL)PENTANOYL-((S)-(-)-(1-NAPHTHYL)ETHYL)AMIDE, MANGANESE (II) ION, Methionine aminopeptidase 2 | | 著者 | Sheppard, G.S, Wang, J, Kawai, M, BaMaung, N.Y, Craig, R.A, Erickson, S.A, Lynch, L, Patel, J, Yang, F, Searle, X.B, Lou, P, Park, C, Kim, K.H, Henkin, J, Lesniewski, R. | | 登録日 | 2003-10-10 | | 公開日 | 2004-10-12 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | 3-Amino-2-hydroxyamides and related compounds as inhibitors of methionine aminopeptidase-2.
Bioorg.Med.Chem.Lett., 14, 2004
|
|
1REF
 
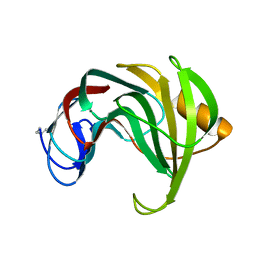 | | ENDO-1,4-BETA-XYLANASE II COMPLEX WITH 2,3-EPOXYPROPYL-BETA-D-XYLOSIDE | | 分子名称: | (2R)-oxiran-2-ylmethyl beta-D-xylopyranoside, BENZOIC ACID, ENDO-1,4-BETA-XYLANASE II | | 著者 | Rouvinen, J, Havukainen, R, Torronen, A. | | 登録日 | 1995-12-21 | | 公開日 | 1997-01-11 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Covalent binding of three epoxyalkyl xylosides to the active site of endo-1,4-xylanase II from Trichoderma reesei.
Biochemistry, 35, 1996
|
|
3M1W
 
 | |
3M29
 
 | |
3M2L
 
 | |
3MMU
 
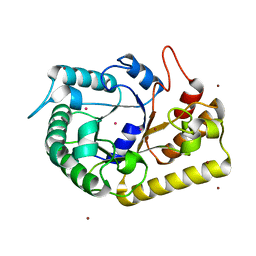 | | Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima | | 分子名称: | CADMIUM ION, Endoglucanase, NICKEL (II) ION | | 著者 | Pereira, J.H, Chen, Z, McAndrew, R.P, Sapra, R, Chhabra, S.R, Sale, K.L, Simmons, B.A, Adams, P.D. | | 登録日 | 2010-04-20 | | 公開日 | 2010-07-28 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.201 Å) | | 主引用文献 | Biochemical characterization and crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima.
J.Struct.Biol., 172, 2010
|
|
3MQ1
 
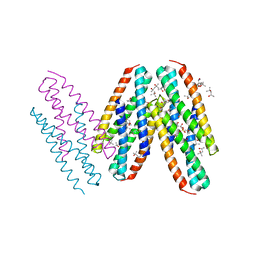 | | Crystal Structure of Dust Mite Allergen Der p 5 | | 分子名称: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, Mite allergen Der p 5, ... | | 著者 | Mueller, G.A, Gosavi, R.A, Krahn, J.M, Edwards, L.L, Cuneo, M.J, Glesner, J, Pomes, A, Chapman, M.D, London, R.E, Pedersen, L.C. | | 登録日 | 2010-04-27 | | 公開日 | 2010-06-02 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Der p 5 crystal structure provides insight into the group 5 dust mite allergens.
J.Biol.Chem., 285, 2010
|
|
7CKO
 
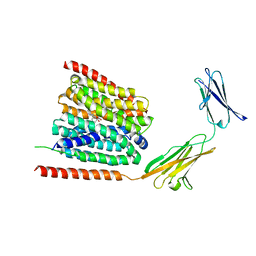 | | Cryo-EM structure of the human MCT1/Basigin-2 complex in the presence of anti-cancer drug candidate 7ACC2 in the inward-open conformation | | 分子名称: | 7-[methyl-(phenylmethyl)amino]-2-oxidanylidene-chromene-3-carboxylic acid, Basigin, Monocarboxylate transporter 1 | | 著者 | Wang, N, Jiang, X, Zhang, S, Zhu, A, Yuan, Y, Lei, J, Yan, C. | | 登録日 | 2020-07-18 | | 公開日 | 2020-12-23 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (2.95 Å) | | 主引用文献 | Structural basis of human monocarboxylate transporter 1 inhibition by anti-cancer drug candidates.
Cell, 184, 2021
|
|
1ROO
 
 | | NMR SOLUTION STRUCTURE OF SHK TOXIN, NMR, 20 STRUCTURES | | 分子名称: | SHK TOXIN | | 著者 | Tudor, J.E, Pallaghy, P.K, Pennington, M.W, Norton, R.S. | | 登録日 | 1996-01-11 | | 公開日 | 1997-01-27 | | 最終更新日 | 2017-11-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of ShK toxin, a novel potassium channel inhibitor from a sea anemone.
Nat.Struct.Biol., 3, 1996
|
|
3M93
 
 | |
3M9A
 
 | |
3M9V
 
 | |
3MS9
 
 | | ABL kinase in complex with imatinib and a fragment (FRAG1) in the myristate pocket | | 分子名称: | 4-(4-METHYL-PIPERAZIN-1-YLMETHYL)-N-[4-METHYL-3-(4-PYRIDIN-3-YL-PYRIMIDIN-2-YLAMINO)-PHENYL]-BENZAMIDE, CHLORIDE ION, Tyrosine-protein kinase ABL1, ... | | 著者 | Cowan-Jacob, S.W, Rummel, G, Fendrich, G. | | 登録日 | 2010-04-29 | | 公開日 | 2010-05-26 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Binding or bending: distinction of allosteric Abl kinase agonists from antagonists by an NMR-based conformational assay.
J.Am.Chem.Soc., 132, 2010
|
|
1RBG
 
 | |
3MT6
 
 | | Structure of ClpP from Escherichia coli in complex with ADEP1 | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACYLDEPSIPEPTIDE 1, ATP-dependent Clp protease proteolytic subunit | | 著者 | Chung, Y.S. | | 登録日 | 2010-04-30 | | 公開日 | 2010-11-03 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.901 Å) | | 主引用文献 | Acyldepsipeptide antibiotics induce the formation of a structured axial channel in ClpP: A model for the ClpX/ClpA-bound state of ClpP.
Chem.Biol., 17, 2010
|
|
3LZZ
 
 | | Crystal structures of Cupin superfamily BbDUF985 from Branchiostoma belcheri tsingtauense in apo and GDP-bound forms | | 分子名称: | ACETATE ION, GUANOSINE-5'-DIPHOSPHATE, Putative uncharacterized protein | | 著者 | Du, Y, He, Y.-X, Saren, G, Zhang, X, Zhang, S.-C, Chen, Y, Zhou, C.-Z. | | 登録日 | 2010-03-02 | | 公開日 | 2010-06-02 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structures of the apo and GDP-bound forms of a cupin-like protein BbDUF985 from Branchiostoma belcheri tsingtauense
Proteins, 2010
|
|
3ME2
 
 | | Crystal structure of mouse RANKL-RANK complex | | 分子名称: | CHLORIDE ION, SODIUM ION, Tumor necrosis factor ligand superfamily member 11, ... | | 著者 | Walter, S.W, Liu, C.Z, Zhu, X.K, Wu, Y, Owens, R.J, Stuart, D.I, Gao, B, Ren, J. | | 登録日 | 2010-03-31 | | 公開日 | 2010-06-02 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural and Functional Insights of RANKL-RANK Interaction and Signaling.
J.Immunol., 2010
|
|
3MF5
 
 | |
1RED
 
 | | ENDO-1,4-BETA-XYLANASE II COMPLEX WITH 4,5-EPOXYPENTYL-BETA-D-XYLOSIDE | | 分子名称: | 3-[(2R)-oxiran-2-yl]propyl beta-D-xylopyranoside, BENZOIC ACID, ENDO-1,4-BETA-XYLANASE II | | 著者 | Rouvinen, J, Havukainen, R, Torronen, A. | | 登録日 | 1995-12-21 | | 公開日 | 1997-01-11 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Covalent binding of three epoxyalkyl xylosides to the active site of endo-1,4-xylanase II from Trichoderma reesei.
Biochemistry, 35, 1996
|
|
