7OMR
 
 | |
7P1N
 
 | | Crystal structure of human acetylcholinesterase in complex with (2R,3R,4S,5S,6R)-2-{4-[1-(4-{5-hydroxy-6-[(E)-(hydroxyimino)methyl]pyridin-2-yl}butyl)-1H-1,2,3-triazol-4-yl]butoxy}-6-(hydroxymethyl)oxane-3,4,5-triol oxime | | 分子名称: | (2R,3R,4S,5S,6R)-2-[4-[1-[4-[6-[(Z)-hydroxyiminomethyl]-5-oxidanyl-pyridin-2-yl]butyl]-1,2,3-triazol-4-yl]butoxy]-6-(hydroxymethyl)oxane-3,4,5-triol, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase, ... | | 著者 | Da Silva, O, Dias, J, Nachon, F. | | 登録日 | 2021-07-02 | | 公開日 | 2022-06-08 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.95 Å) | | 主引用文献 | A New Class of Bi- and Trifunctional Sugar Oximes as Antidotes against Organophosphorus Poisoning.
J.Med.Chem., 65, 2022
|
|
7OMD
 
 | | Crystal structure of azacoelenterazine-bound Renilla reniformis luciferase variant RLuc8-D162A | | 分子名称: | 6-(4-hydroxyphenyl)-2-[(4-hydroxyphenyl)methyl]-8-(phenylmethyl)-[1,2,4]triazolo[4,3-a]pyrazin-3-one, CHLORIDE ION, Coelenterazine h 2-monooxygenase, ... | | 著者 | Schenkmayerova, A, Janin, Y.L, Marek, M. | | 登録日 | 2021-05-21 | | 公開日 | 2022-06-01 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.601 Å) | | 主引用文献 | Catalytic mechanism for Renilla-type luciferases
Nat Catal, 2023
|
|
6XT1
 
 | |
7OVX
 
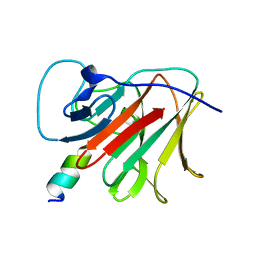 | | E3 RING ligase binding domain | | 分子名称: | E3 ubiquitin-protein ligase TRIM7, Peptide G | | 著者 | James, L.C. | | 登録日 | 2021-06-15 | | 公開日 | 2022-06-22 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | E3 ligase targeting domain
To Be Published
|
|
4U74
 
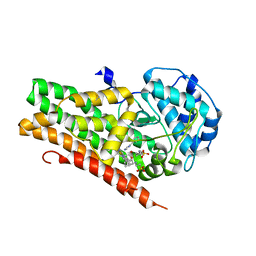 | | Crystal structure of 4-phenylimidazole bound form of human indoleamine 2,3-dioxygenase (G262A mutant) | | 分子名称: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, 4-PHENYL-1H-IMIDAZOLE, Indoleamine 2,3-dioxygenase 1, ... | | 著者 | Sugimoto, H, Horitani, M, Kometani, E, Shiro, Y. | | 登録日 | 2014-07-30 | | 公開日 | 2015-09-02 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.31 Å) | | 主引用文献 | Conformation and Mobility of Active Site Loop is Critical for Substrate Binding and Inhibition in Human Indoleamine 2,3-Dioxygenase
to be published
|
|
7P80
 
 | | Crystal structure of ClpP from Bacillus subtilis in complex with ADEP2 (compressed state) | | 分子名称: | ADEP2, ATP-dependent Clp protease proteolytic subunit | | 著者 | Lee, B.-G, Kim, L, Kim, M.K, Kwon, D.H, Song, H.K. | | 登録日 | 2021-07-21 | | 公開日 | 2022-06-29 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.98 Å) | | 主引用文献 | Structural insights into ClpP protease side exit pore-opening by a pH drop coupled with substrate hydrolysis.
Embo J., 41, 2022
|
|
7P81
 
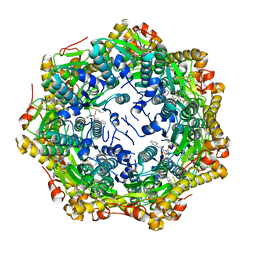 | | Crystal structure of ClpP from Bacillus subtilis in complex with ADEP2 (compact state) | | 分子名称: | ADEP2, ATP-dependent Clp protease proteolytic subunit | | 著者 | Lee, B.-G, Kim, L, Kim, M.K, Kwon, D.H, Song, H.K. | | 登録日 | 2021-07-21 | | 公開日 | 2022-06-29 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.79 Å) | | 主引用文献 | Structural insights into ClpP protease side exit pore-opening by a pH drop coupled with substrate hydrolysis.
Embo J., 41, 2022
|
|
7P4E
 
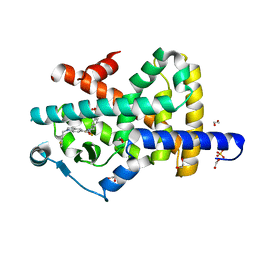 | | Crystal structure of PPARgamma in complex with compound FL217 | | 分子名称: | 1,2-ETHANEDIOL, Peroxisome proliferator-activated receptor gamma, SULFATE ION, ... | | 著者 | Ni, X, Lillich, F, Proschak, E, Chaikuad, A, Knapp, S, Structural Genomics Consortium (SGC) | | 登録日 | 2021-07-11 | | 公開日 | 2022-07-06 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structure-Based Design of Dual Partial Peroxisome Proliferator-Activated Receptor gamma Agonists/Soluble Epoxide Hydrolase Inhibitors.
J.Med.Chem., 64, 2021
|
|
7OJ1
 
 | |
7OW2
 
 | | E3 RING ligase binding domain with peptide | | 分子名称: | CHLORIDE ION, E3 ubiquitin-protein ligase RNF187 peptide, E3 ubiquitin-protein ligase TRIM7, ... | | 著者 | James, L.C. | | 登録日 | 2021-06-16 | | 公開日 | 2022-07-13 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.17 Å) | | 主引用文献 | E3 ligase targeting domain
To Be Published
|
|
7P0W
 
 | | Crystal structure of a trapped Pab-AGOG/double-standed DNA covalent intermediate (DNA containing thymine opposite to lesion) | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(*AP*GP*AP*AP*AP*TP*AP*AP*A)-3'), DNA (5'-D(*TP*TP*TP*(PED)P*TP*TP*TP*CP*T)-3'), ... | | 著者 | Coste, F, Goffinont, S, Flament, D, Castaing, B. | | 登録日 | 2021-06-30 | | 公開日 | 2022-07-13 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.12 Å) | | 主引用文献 | Structural and functional determinants of the archaeal 8-oxoguanine-DNA glycosylase AGOG for DNA damage recognition and processing.
Nucleic Acids Res., 50, 2022
|
|
7OXE
 
 | |
7OJU
 
 | | Chaetomium thermophilum Naa50 GNAT-domain in complex with bisubstrate analogue CoA-Ac-MVNAL | | 分子名称: | CARBOXYMETHYL COENZYME *A, GLYCEROL, HEXAETHYLENE GLYCOL, ... | | 著者 | Weidenhausen, J, Kopp, J, Sinning, I. | | 登録日 | 2021-05-17 | | 公開日 | 2022-09-28 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Extended N-Terminal Acetyltransferase Naa50 in Filamentous Fungi Adds to Naa50 Diversity.
Int J Mol Sci, 23, 2022
|
|
7OYE
 
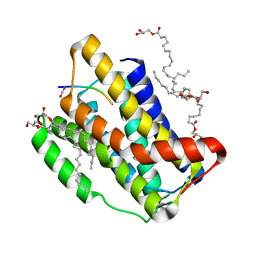 | |
6XQF
 
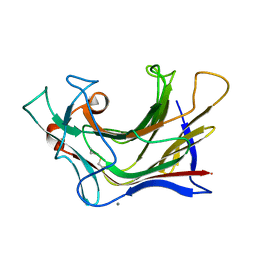 | | Crystal structure of SCLam E144S mutant, a non-specific endo-beta-1,3(4)-glucanase from family GH16, co-crystallized with 1,3-beta-D-cellotriosyl-glucose, presenting a 1,3-beta-D-cellobiosyl-glucose at active site | | 分子名称: | CALCIUM ION, GH16 family protein, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-3)-alpha-D-glucopyranose | | 著者 | Liberato, M.V, Squina, F. | | 登録日 | 2020-07-09 | | 公開日 | 2021-02-10 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.58 Å) | | 主引用文献 | Insights into the dual cleavage activity of the GH16 laminarinase enzyme class on beta-1,3 and beta-1,4 glycosidic bonds.
J.Biol.Chem., 296, 2021
|
|
7P5T
 
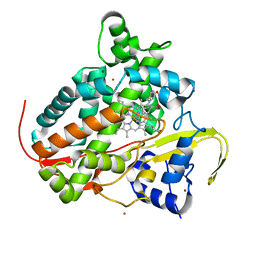 | | Structure of CYP142 from Mycobacterium tuberculosis in complex with inhibitor MEK216 | | 分子名称: | BROMIDE ION, POTASSIUM ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | 著者 | Snee, M, Kavanagh, M, Tunnicliffe, R, McLean, K, Levy, C, Munro, A. | | 登録日 | 2021-07-14 | | 公開日 | 2022-11-16 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Structure of CYP142 from Mycobacterium tuberculosis in complex with inhibitor MEK216
To Be Published
|
|
7ONG
 
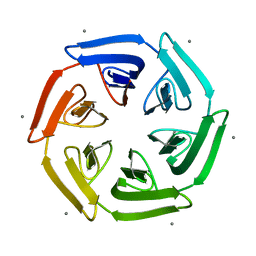 | | Crystal structure of the computationally designed SAKe6BE-L1 protein | | 分子名称: | CALCIUM ION, SAKe6BE-L1 | | 著者 | Wouters, S.M.L, Noguchi, H, Velpula, G, Clarke, D.E, Voet, A.R.D, De Feyter, S. | | 登録日 | 2021-05-25 | | 公開日 | 2022-12-14 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | SAKe: Computationally Designed Modular Protein Building Blocks for Macromolecular Assemblies
To be published
|
|
7ON8
 
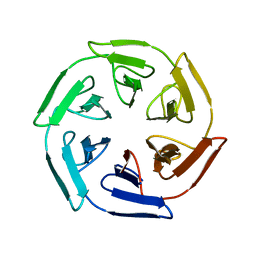 | |
7OPU
 
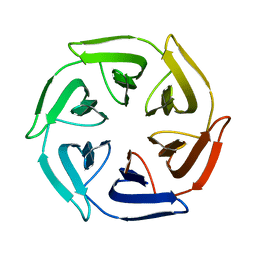 | |
7ONA
 
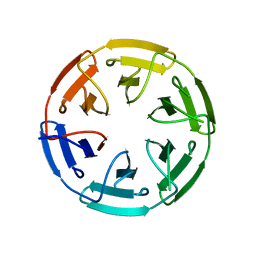 | | Crystal structure of the computationally designed SAKe6AC protein | | 分子名称: | CALCIUM ION, SAKe6AC | | 著者 | Wouters, S.M.L, Noguchi, H, Velpula, G, Clarke, D.E, Voet, A.R.D, De Feyter, S. | | 登録日 | 2021-05-25 | | 公開日 | 2022-12-14 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | SAKe: Computationally Designed Modular Protein Building Blocks for Macromolecular Assemblies
To be published
|
|
7ON7
 
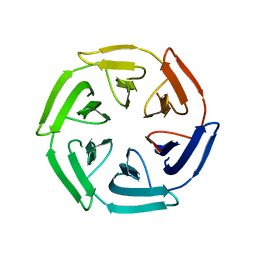 | |
7ONE
 
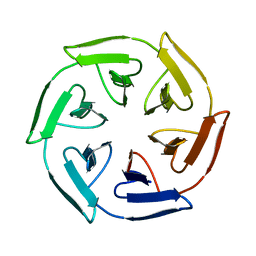 | | Crystal structure of the self-assembled SAKe6BE designer protein | | 分子名称: | SAKe6BE | | 著者 | Wouters, S.M.L, Noguchi, H, Velpula, G, Clarke, D.E, Voet, A.R.D, De Feyter, S. | | 登録日 | 2021-05-25 | | 公開日 | 2022-12-14 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | SAKe: Computationally Designed Modular Protein Building Blocks for Macromolecular Assemblies
To be published
|
|
7ONC
 
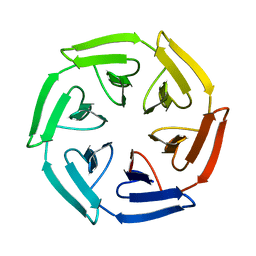 | | Crystal structure of the computationally designed SAKe6BE protein | | 分子名称: | SAKe6BE | | 著者 | Wouters, S.M.L, Noguchi, H, Velpula, G, Clarke, D.E, Voet, A.R.D, De Feyter, S. | | 登録日 | 2021-05-25 | | 公開日 | 2022-12-14 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.49 Å) | | 主引用文献 | SAKe: Computationally Designed Modular Protein Building Blocks for Macromolecular Assemblies
To be published
|
|
7ONH
 
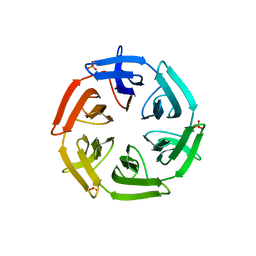 | | Crystal structure of the computationally designed SAKe6BE-L3 protein | | 分子名称: | SAKe6BE-L3, SULFATE ION | | 著者 | Wouters, S.M.L, Noguchi, H, Velpula, G, Clarke, D.E, Voet, A.R.D, De Feyter, S. | | 登録日 | 2021-05-25 | | 公開日 | 2022-12-14 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | SAKe: Computationally Designed Modular Protein Building Blocks for Macromolecular Assemblies
To be published
|
|
