3WX4
 
 | |
3WPQ
 
 | |
3WTJ
 
 | | Crystal Structure of Lymnaea stagnalis Acetylcholine Binding Protein Complexed with Thiacloprid | | 分子名称: | Acetylcholine-binding protein, {(2Z)-3-[(6-chloropyridin-3-yl)methyl]-1,3-thiazolidin-2-ylidene}cyanamide | | 著者 | Okajima, T, Ihara, M, Yamashita, A, Oda, T, Matsuda, K. | | 登録日 | 2014-04-11 | | 公開日 | 2015-02-04 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.24 Å) | | 主引用文献 | Studies on an acetylcholine binding protein identify a basic residue in loop G on the beta 1 strand as a new structural determinant of neonicotinoid actions
Mol.Pharmacol., 86, 2014
|
|
3WHP
 
 | | Crystal structure of the C-terminal domain of Themus thermophilus LitR in complex with cobalamin | | 分子名称: | COBALAMIN, Probable transcriptional regulator | | 著者 | Agari, Y, Takano, H, Beppu, T, Ueda, K, Shinkai, A. | | 登録日 | 2013-08-30 | | 公開日 | 2014-09-03 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.52 Å) | | 主引用文献 | Crystal structure of the C-terminal domain of Themus thermophilus LitR in complex with cobalamin
To be Published
|
|
3WTK
 
 | | Crystal Structure of Lymnaea stagnalis Acetylcholine-Binding Protein Q55R Mutant Complexed with Thiacloprid | | 分子名称: | Acetylcholine-binding protein, {(2Z)-3-[(6-chloropyridin-3-yl)methyl]-1,3-thiazolidin-2-ylidene}cyanamide | | 著者 | Okajima, T, Ihara, M, Yamashita, A, Oda, T, Matsuda, K. | | 登録日 | 2014-04-11 | | 公開日 | 2015-02-04 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.69 Å) | | 主引用文献 | Studies on an acetylcholine binding protein identify a basic residue in loop G on the beta 1 strand as a new structural determinant of neonicotinoid actions
Mol.Pharmacol., 86, 2014
|
|
7QIJ
 
 | |
3WTH
 
 | | Crystal Structure of Lymnaea stagnalis Acetylcholine-Binding Protein Q55R Mutant Complexed with Imidacloprid | | 分子名称: | (2E)-1-[(6-chloropyridin-3-yl)methyl]-N-nitroimidazolidin-2-imine, Acetylcholine-binding protein | | 著者 | Okajima, T, Ihara, M, Yamashita, A, Oda, T, Matsuda, K. | | 登録日 | 2014-04-11 | | 公開日 | 2015-02-04 | | 実験手法 | X-RAY DIFFRACTION (2.54 Å) | | 主引用文献 | Studies on an acetylcholine binding protein identify a basic residue in loop G on the beta 1 strand as a new structural determinant of neonicotinoid actions
Mol.Pharmacol., 86, 2014
|
|
3OKZ
 
 | | Crystal Structure of protein gbs0355 from Streptococcus agalactiae, Northeast Structural Genomics Consortium Target SaR127 | | 分子名称: | Putative uncharacterized protein gbs0355 | | 著者 | Kuzin, A, Lew, S, Vorobiev, S.M, Seetharaman, J, Janjua, J, Xiao, R, Ciccosanti, C, Wang, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2010-08-25 | | 公開日 | 2010-09-22 | | 最終更新日 | 2019-07-17 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Northeast Structural Genomics Consortium Target SaR127
To be Published
|
|
3WQH
 
 | | Crystal Structure of human DPP-IV in complex with Anagliptin | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase 4, N-[2-({2-[(2S)-2-cyanopyrrolidin-1-yl]-2-oxoethyl}amino)-2-methylpropyl]-2-methylpyrazolo[1,5-a]pyrimidine-6-carboxamide | | 著者 | Watanabe, Y.S, Okada, S, Motoyama, T, Takahashi, R, Adachi, H, Oka, M. | | 登録日 | 2014-01-27 | | 公開日 | 2015-07-15 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | Anagliptin, a potent dipeptidyl peptidase IV inhibitor: its single-crystal structure and enzyme interactions.
J Enzyme Inhib Med Chem, 30, 2015
|
|
7LB7
 
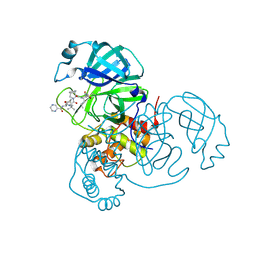 | | Joint X-ray/neutron structure of SARS-CoV-2 main protease (3CL Mpro) in complex with Telaprevir | | 分子名称: | (1S,3aR,6aS)-2-[(2S)-2-({(2S)-2-cyclohexyl-2-[(pyrazin-2-ylcarbonyl)amino]acetyl}amino)-3,3-dimethylbutanoyl]-N-[(2R,3S)-1-(cyclopropylamino)-2-hydroxy-1-oxohexan-3-yl]octahydrocyclopenta[c]pyrrole-1-carboxamide, 3C-like proteinase | | 著者 | Kovalevsky, A.Y, Kneller, D.W, Coates, L. | | 登録日 | 2021-01-07 | | 公開日 | 2021-01-20 | | 最終更新日 | 2024-04-03 | | 実験手法 | NEUTRON DIFFRACTION (2 Å), X-RAY DIFFRACTION | | 主引用文献 | Direct Observation of Protonation State Modulation in SARS-CoV-2 Main Protease upon Inhibitor Binding with Neutron Crystallography.
J.Med.Chem., 64, 2021
|
|
3ZQB
 
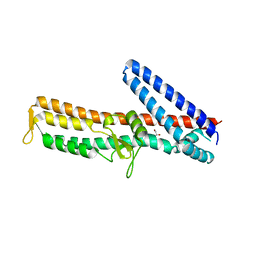 | | PrgI-SipD from Salmonella typhimurium | | 分子名称: | GLYCEROL, PROTEIN PRGI, CELL INVASION PROTEIN SIPD | | 著者 | Lunelli, M, Kolbe, M. | | 登録日 | 2011-06-08 | | 公開日 | 2011-08-17 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal Structure of Prgi-Sipd: Insight Into a Secretion Competent State of the Type Three Secretion System Needle Tip and its Interaction with Host Ligands
Plos Pathog., 7, 2011
|
|
7RK8
 
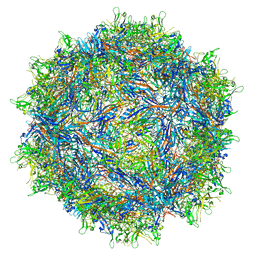 | |
4AHD
 
 | | Q12L - Angiogenin mutants and amyotrophic lateral sclerosis - a biochemical and biological analysis | | 分子名称: | ANGIOGENIN | | 著者 | Thiyagarajan, N, Ferguson, R, Saha, S, Pham, T, Subramanian, V, Acharya, K.R. | | 登録日 | 2012-02-06 | | 公開日 | 2012-10-10 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.47 Å) | | 主引用文献 | Structural and Molecular Insights Into the Mechanism of Action of Human Angiogenin-Als Variants in Neurons.
Nat.Commun., 3, 2012
|
|
7RK9
 
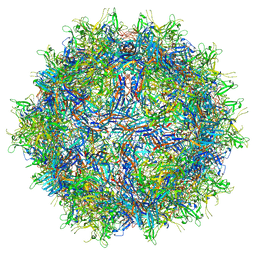 | |
7YAS
 
 | | HYDROXYNITRILE LYASE, LOW TEMPERATURE NATIVE STRUCTURE | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, PROTEIN (HYDROXYNITRILE LYASE), ... | | 著者 | Zuegg, J, Wagner, U.G, Gugganig, M, Kratky, C. | | 登録日 | 1999-03-15 | | 公開日 | 1999-10-13 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Three-dimensional structures of enzyme-substrate complexes of the hydroxynitrile lyase from Hevea brasiliensis.
Protein Sci., 8, 1999
|
|
7RL1
 
 | | AAVrh.10-7x capsid | | 分子名称: | Capsid protein VP1, DNA (5'-D(*CP*A)-3') | | 著者 | Mietzsch, M, McKenna, R. | | 登録日 | 2021-07-23 | | 公開日 | 2021-09-08 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (2.71 Å) | | 主引用文献 | Structural Study of Aavrh.10 Receptor and Antibody Interactions.
J.Virol., 95, 2021
|
|
429D
 
 | |
3MVE
 
 | | Crystal structure of a novel pyruvate decarboxylase | | 分子名称: | 1,2-ETHANEDIOL, SULFATE ION, UPF0255 protein VV1_0328 | | 著者 | Cha, S.S, Jeong, C.S, An, Y.J. | | 登録日 | 2010-05-04 | | 公開日 | 2011-05-18 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | FrsA functions as a cofactor-independent decarboxylase to control metabolic flux.
Nat.Chem.Biol., 7, 2011
|
|
4A4L
 
 | | CRYSTAL STRUCTURE OF POLO-LIKE KINASE 1 IN COMPLEX WITH A 5-(2-AMINO- PYRIMIDIN-4-YL)-1H-PYRROLE INHIBITOR | | 分子名称: | 1-METHYL-5-(2-{[5-(4-METHYLPIPERAZIN-1-YL)-2-(TRIFLUOROMETHOXY)PHENYL]AMINO}PYRIMIDIN-4-YL)-1H-PYRROLE-3-CARBOXAMIDE, L(+)-TARTARIC ACID, SERINE/THREONINE-PROTEIN KINASE PLK1, ... | | 著者 | Bertrand, J.A, Bossi, R.T. | | 登録日 | 2011-10-17 | | 公開日 | 2012-01-11 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | 5-(2-Amino-Pyrimidin-4-Yl)-1H-Pyrrole and 2-(2-Amino-Pyrimidin-4-Yl)-1,5,6,7-Tetrahydro-Pyrrolo[3,2-C]Pyridin-4-One Derivatives as New Classes of Selective and Orally Available Polo-Like Kinase 1 Inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
7LZQ
 
 | | Crystal structure of the BCL6 BTB domain in complex with OICR-4425 | | 分子名称: | B-cell lymphoma 6 protein, DIMETHYL SULFOXIDE, N-(3-chloropyridin-4-yl)-2-(5-methyl-4-oxo-4,5-dihydro-1H-pyrrolo[3,2-c]pyridin-1-yl)acetamide | | 著者 | Kuntz, D.A, Prive, G.G. | | 登録日 | 2021-03-10 | | 公開日 | 2022-08-17 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.71 Å) | | 主引用文献 | Discovery of OICR12694: A Novel, Potent, Selective, and Orally Bioavailable BCL6 BTB Inhibitor.
Acs Med.Chem.Lett., 14, 2023
|
|
7MHT
 
 | | CYTOSINE-SPECIFIC METHYLTRANSFERASE HHAI/DNA COMPLEX | | 分子名称: | 5'-D(P*CP*CP*AP*TP*GP*AP*GP*CP*TP*GP*AP*C)-3', 5'-D(P*GP*TP*CP*AP*GP*CP*GP*CP*AP*TP*GP*G)-3', CYTOSINE-SPECIFIC METHYLTRANSFERASE HHAI, ... | | 著者 | O'Gara, M, Horton, J.R, Roberts, R.J, Cheng, X. | | 登録日 | 1998-08-05 | | 公開日 | 1998-11-30 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.87 Å) | | 主引用文献 | Structures of HhaI methyltransferase complexed with substrates containing mismatches at the target base.
Nat.Struct.Biol., 5, 1998
|
|
7MDH
 
 | | STRUCTURAL BASIS FOR LIGHT ACITVATION OF A CHLOROPLAST ENZYME. THE STRUCTURE OF SORGHUM NADP-MALATE DEHYDROGENASE IN ITS OXIDIZED FORM | | 分子名称: | PROTEIN (MALATE DEHYDROGENASE), ZINC ION | | 著者 | Johansson, K, Ramaswamy, S, Saarinen, M, Lemaire-Chamley, M, Issakidis-Bourguet, E, Miginiac-Maslow, M, Eklund, H. | | 登録日 | 1999-02-16 | | 公開日 | 1999-06-04 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural basis for light activation of a chloroplast enzyme: the structure of sorghum NADP-malate dehydrogenase in its oxidized form.
Biochemistry, 38, 1999
|
|
474D
 
 | | A NOVEL END-TO-END BINDING OF TWO NETROPSINS TO THE DNA DECAMER D(CCCCCIIIII)2 | | 分子名称: | DNA (5'-D(*CP*CP*CP*(CBR)P*CP*IP*IP*IP*IP*I)-3'), NETROPSIN | | 著者 | Chen, X, Rao, S.T, Sekar, K, Sundaralingam, M. | | 登録日 | 1998-01-14 | | 公開日 | 1998-12-02 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | A novel end-to-end binding of two netropsins to the DNA decamers d(CCCCCIIIII)2, d(CCCBr5CCIIIII)2and d(CBr5CCCCIIIII)2.
Nucleic Acids Res., 26, 1998
|
|
462D
 
 | | CRYSTAL STRUCTURE OF THE HIV-1 GENOMIC RNA DIMERIZATION INITIATION SITE | | 分子名称: | MAGNESIUM ION, RNA (5'-R(*CP*UP*UP*GP*CP*UP*GP*AP*GP*GP*UP*GP*CP*AP*CP*AP*CP*AP*GP*CP*AP*AP*G) -3') | | 著者 | Ennifar, E, Yusupov, M, Walter, P, Marquet, R, Ehresmann, C, Ehresmann, B, Dumas, P. | | 登録日 | 1999-03-18 | | 公開日 | 1999-12-02 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | The crystal structure of the dimerization initiation site of genomic HIV-1 RNA reveals an extended duplex with two adenine bulges.
Structure Fold.Des., 7, 1999
|
|
472D
 
 | | STRUCTURE OF AN OCTAMER RNA WITH TANDEM GG/UU MISPAIRS | | 分子名称: | RNA (5'-R(*GP*UP*AP*GP*GP*CP*AP*C)-3'), RNA (5'-R(*GP*UP*GP*UP*UP*UP*AP*C)-3') | | 著者 | Deng, J, Sundaralingam, M. | | 登録日 | 1999-05-14 | | 公開日 | 2000-11-27 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Synthesis and crystal structure of an octamer RNA r(guguuuac)/r(guaggcac) with G.G/U.U tandem wobble base pairs: comparison with other tandem G.U pairs.
Nucleic Acids Res., 28, 2000
|
|
