6T36
 
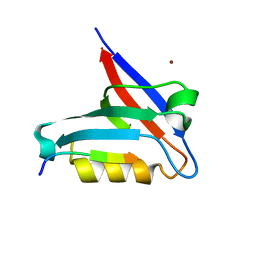 | | Crystal structure of the PTPN3 PDZ domain bound to the HBV core protein C-terminal peptide | | 分子名称: | BROMIDE ION, Capsid protein, Tyrosine-protein phosphatase non-receptor type 3 | | 著者 | Genera, M, Mechaly, A, Haouz, A, Caillet-Saguy, C. | | 登録日 | 2019-10-10 | | 公開日 | 2021-01-20 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.86 Å) | | 主引用文献 | Molecular basis of the interaction of the human tyrosine phosphatase PTPN3 with the hepatitis B virus core protein.
Sci Rep, 11, 2021
|
|
7NRD
 
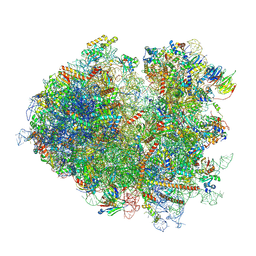 | | Structure of the yeast Gcn1 bound to a colliding stalled 80S ribosome with MBF1, A/P-tRNA and P/E-tRNA | | 分子名称: | 25S rRNA (3184-MER), 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, ... | | 著者 | Pochopien, A.A, Beckert, B, Wilson, D.N. | | 登録日 | 2021-03-03 | | 公開日 | 2021-04-14 | | 最終更新日 | 2025-07-09 | | 実験手法 | ELECTRON MICROSCOPY (4.36 Å) | | 主引用文献 | Structure of Gcn1 bound to stalled and colliding 80S ribosomes.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7NRC
 
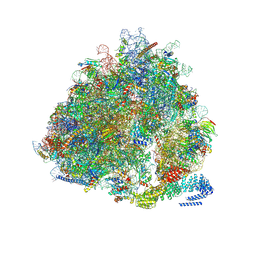 | | Structure of the yeast Gcn1 bound to a leading stalled 80S ribosome with Rbg2, Gir2, A- and P-tRNA and eIF5A | | 分子名称: | 18S rRNA (1771-MER), 25S rRNA (3184-MER), 40S ribosomal protein S0-A, ... | | 著者 | Pochopien, A.A, Beckert, B, Wilson, D.N. | | 登録日 | 2021-03-03 | | 公開日 | 2021-05-05 | | 最終更新日 | 2025-04-09 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Structure of Gcn1 bound to stalled and colliding 80S ribosomes.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
2I9K
 
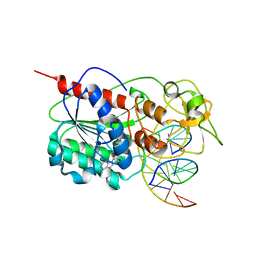 | | Engineered Extrahelical Base Destabilization Enhances Sequence Discrimination of DNA Methyltransferase M.HhaI | | 分子名称: | 5'-D(*T*GP*AP*TP*AP*GP*CP*GP*CP*TP*AP*TP*C)-3', Modification methylase HhaI, S-ADENOSYL-L-HOMOCYSTEINE | | 著者 | Youngblood, B, Shieh, F.K, De Los Rios, S, Perona, J.J, Reich, N.O. | | 登録日 | 2006-09-05 | | 公開日 | 2006-10-10 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Engineered Extrahelical Base Destabilization Enhances Sequence Discrimination of DNA Methyltransferase M.HhaI
J.Mol.Biol., 362, 2006
|
|
5DGF
 
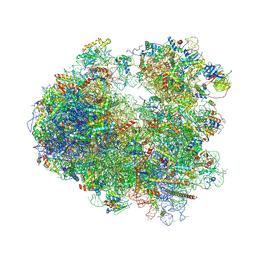 | | Complex of yeast 80S ribosome with hypusine-containing/non-modified eIF5A and/or a peptidyl-tRNA analog | | 分子名称: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | 著者 | Melnikov, S, Mailliot, J, Shin, B.-S, Rigger, L, Yusupova, G, Micura, R, Dever, T.E, Yusupov, M. | | 登録日 | 2015-08-27 | | 公開日 | 2016-12-14 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Coping with proline stalling: structural basis of hypusine-induced protein synthesis by the eukaryotic ribosome
To Be Published
|
|
4D1C
 
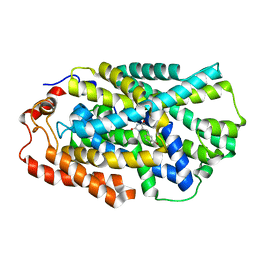 | | STRUCTURE OF MHP1, A NUCLEOBASE-CATION-SYMPORT-1 FAMILY TRANSPORTER, IN A CLOSED CONFORMATION WITH bromovinylhydantoin bound. | | 分子名称: | (5Z)-5-[(3-bromophenyl)methylidene]imidazolidine-2,4-dione, HYDANTOIN TRANSPORT PROTEIN, SODIUM ION | | 著者 | Weyand, S, Brueckner, F, Geng, T, Drew, D, Iwata, S, Henderson, P.J.F, Cameron, A.D. | | 登録日 | 2014-05-01 | | 公開日 | 2014-07-02 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3.7 Å) | | 主引用文献 | Molecular Mechanism of Ligand Recognition by Membrane Transport Protein, Mhp1.
Embo J., 33, 2014
|
|
5DGE
 
 | | Coping with proline stalling: structural basis of hypusine-induced protein synthesis by the eukaryotic ribosome | | 分子名称: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | 著者 | Melnikov, S, Mailliot, J, Shin, B.-S, Rigger, L, Yusupova, G, Micura, R, Dever, T.E, Yusupov, M. | | 登録日 | 2015-08-27 | | 公開日 | 2017-01-25 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (3.45 Å) | | 主引用文献 | Coping with proline stalling: structural basis of hypusine-induced protein synthesis by the eukaryotic ribosome
To Be Published
|
|
8RXH
 
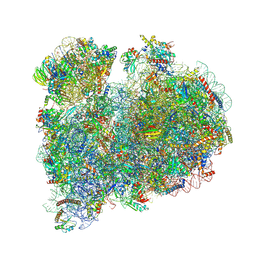 | | CRYO-EM STRUCTURE OF LEISHMANIA MAJOR 80S RIBOSOME WITH A/P/E-site tRNA AND mRNA : PARENTAL STRAIN | | 分子名称: | (2S)-2-[2-[4-[[(2R,3S,4S)-3-acetyloxy-4-oxidanyl-pyrrolidin-2-yl]methyl]phenoxy]ethanoylamino]-6-azanyl-hexanoic acid, 40S ribosomal protein S12, 40S ribosomal protein S14, ... | | 著者 | Rajan, K.S, Yonath, A. | | 登録日 | 2024-02-07 | | 公開日 | 2024-05-15 | | 最終更新日 | 2024-11-20 | | 実験手法 | ELECTRON MICROSCOPY (2.93 Å) | | 主引用文献 | Structural and mechanistic insights into the function of Leishmania ribosome lacking a single pseudouridine modification.
Cell Rep, 43, 2024
|
|
8RXX
 
 | |
6Z6L
 
 | | Cryo-EM structure of human CCDC124 bound to 80S ribosomes | | 分子名称: | 18S rRNA, 28S rRNA, 40S ribosomal protein S10, ... | | 著者 | Wells, J.N, Buschauer, R, Mackens-Kiani, T, Best, K, Kratzat, H, Berninghausen, O, Becker, T, Cheng, J, Beckmann, R. | | 登録日 | 2020-05-28 | | 公開日 | 2020-07-29 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structure and function of yeast Lso2 and human CCDC124 bound to hibernating ribosomes.
Plos Biol., 18, 2020
|
|
5DAT
 
 | | Complex of yeast 80S ribosome with hypusine-containing eIF5A | | 分子名称: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | 著者 | Melnikov, S, Mailliot, J, Shin, B.-S, Rigger, L, Yusupova, G, Micura, R, Dever, T.E, Yusupov, M. | | 登録日 | 2015-08-20 | | 公開日 | 2016-08-31 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (3.15 Å) | | 主引用文献 | Coping with proline stalling: structural basis of hypusine-induced protein synthesis by the eukaryotic ribosome
To Be Published
|
|
4D1D
 
 | | STRUCTURE OF MHP1, A NUCLEOBASE-CATION-SYMPORT-1 FAMILY TRANSPORTER with the inhibitor 5-(2-naphthylmethyl)-L-hydantoin. | | 分子名称: | 5-(2-NAPHTHYLMETHYL)-D-HYDANTOIN, 5-(2-NAPHTHYLMETHYL)-L-HYDANTOIN, HYDANTOIN TRANSPORT PROTEIN, ... | | 著者 | Weyand, S, Brueckner, F, Geng, T, Drew, D, Iwata, S, Henderson, P.J.F, Cameron, A.D. | | 登録日 | 2014-05-01 | | 公開日 | 2014-07-02 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3.7 Å) | | 主引用文献 | Molecular Mechanism of Ligand Recognition by Membrane Transport Protein, Mhp1.
Embo J., 33, 2014
|
|
4D1B
 
 | | STRUCTURE OF MHP1, A NUCLEOBASE-CATION-SYMPORT-1 FAMILY TRANSPORTER, IN A CLOSED CONFORMATION WITH BENZYL-HYDANTOIN | | 分子名称: | (5S)-5-benzylimidazolidine-2,4-dione, HYDANTOIN TRANSPORT PROTEIN, SODIUM ION | | 著者 | Brueckner, F, Geng, T, Weyand, S, Drew, D, Iwata, S, Henderson, P.J.F, Cameron, A.D. | | 登録日 | 2014-05-01 | | 公開日 | 2014-07-02 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3.8 Å) | | 主引用文献 | Molecular Mechanism of Ligand Recognition by Membrane Transport Protein, Mhp1.
Embo J., 33, 2014
|
|
4D1A
 
 | | STRUCTURE OF MHP1, A NUCLEOBASE-CATION-SYMPORT-1 FAMILY TRANSPORTER, IN A CLOSED CONFORMATION WITH INDOLYLMETHYL-HYDANTOIN | | 分子名称: | (5S)-5-(1H-indol-3-ylmethyl)imidazolidine-2,4-dione, HYDANTOIN TRANSPORT PROTEIN, SODIUM ION | | 著者 | Weyand, S, Brueckner, F, Geng, T, Drew, D, Iwata, S, Henderson, P.J.F, Cameron, A.D. | | 登録日 | 2014-05-01 | | 公開日 | 2014-07-02 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | Molecular Mechanism of Ligand Recognition by Membrane Transport Protein, Mhp1.
Embo J., 33, 2014
|
|
8RJ6
 
 | | E. coli adenylate kinase in complex with ATP and AMP and Mg2+ as a result of enzymatic AP4A hydrolysis. | | 分子名称: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Tischlik, S, Ronge, P, Wolf-Watz, M, Sauer-Eriksson, A.E. | | 登録日 | 2023-12-20 | | 公開日 | 2024-07-10 | | 最終更新日 | 2024-08-28 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Magnesium induced structural reorganization in the active site of adenylate kinase.
Sci Adv, 10, 2024
|
|
8RJ4
 
 | | E. coli adenylate kinase in complex with two ADP molecules and Mg2+ as a result of enzymatic AP4A hydrolysis | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Adenylate kinase, CHLORIDE ION, ... | | 著者 | Tischlik, S, Ronge, P, Wolf-Watz, M, Sauer-Eriksson, A.E. | | 登録日 | 2023-12-20 | | 公開日 | 2024-07-10 | | 最終更新日 | 2024-08-28 | | 実験手法 | X-RAY DIFFRACTION (2.11 Å) | | 主引用文献 | Magnesium induced structural reorganization in the active site of adenylate kinase.
Sci Adv, 10, 2024
|
|
6ZAY
 
 | |
5DC3
 
 | | Complex of yeast 80S ribosome with non-modified eIF5A | | 分子名称: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | 著者 | Melnikov, S, Mailliot, J, Shin, B.-S, Rigger, L, Yusupova, G, Micura, R, Dever, T.E, Yusupov, M. | | 登録日 | 2015-08-23 | | 公開日 | 2016-06-01 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (3.25 Å) | | 主引用文献 | Crystal Structure of Hypusine-Containing Translation Factor eIF5A Bound to a Rotated Eukaryotic Ribosome.
J.Mol.Biol., 428, 2016
|
|
7FC2
 
 | | Crystal Structure of GPX6 | | 分子名称: | Glutathione peroxidase 6, SULFATE ION | | 著者 | Sun, J. | | 登録日 | 2021-07-13 | | 公開日 | 2022-07-20 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal Structure of GPX6
To Be Published
|
|
4U53
 
 | | Crystal structure of Deoxynivalenol bound to the yeast 80S ribosome | | 分子名称: | (3beta,7alpha)-3,7,15-trihydroxy-12,13-epoxytrichothec-9-en-8-one, 18S ribosomal RNA, 25S ribosomal RNA, ... | | 著者 | Garreau de Loubresse, N, Prokhorova, I, Yusupova, G, Yusupov, M. | | 登録日 | 2014-07-24 | | 公開日 | 2014-10-22 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Structural basis for the inhibition of the eukaryotic ribosome.
Nature, 513, 2014
|
|
9L14
 
 | | Crystal structure of the monobody CL-1 in complex with the Escherichia coli adenylate kinase | | 分子名称: | Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Nakamura, I, Amesaka, H, Tanaka, S.I, Matsuo, T. | | 登録日 | 2024-12-13 | | 公開日 | 2025-06-04 | | 実験手法 | X-RAY DIFFRACTION (1.86 Å) | | 主引用文献 | Binding mechanism of adenylate kinase-specific monobodies.
Febs Lett., 2025
|
|
6EML
 
 | | Cryo-EM structure of a late pre-40S ribosomal subunit from Saccharomyces cerevisiae | | 分子名称: | 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, 40S ribosomal protein S11-A, ... | | 著者 | Heuer, A, Thomson, E, Schmidt, C, Berninghausen, O, Becker, T, Hurt, E, Beckmann, R. | | 登録日 | 2017-10-02 | | 公開日 | 2017-11-29 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Cryo-EM structure of a late pre-40S ribosomal subunit fromSaccharomyces cerevisiae.
Elife, 6, 2017
|
|
4U4R
 
 | | Crystal structure of Lactimidomycin bound to the yeast 80S ribosome | | 分子名称: | 18S ribosomal RNA, 25S ribosomal RNA, 4-{(2R,5S,6E)-2-hydroxy-5-methyl-7-[(2R,3S,4E,6Z,10E)-3-methyl-12-oxooxacyclododeca-4,6,10-trien-2-yl]-4-oxooct-6-en-1-yl}piperidine-2,6-dione, ... | | 著者 | Garreau de Loubresse, N, Prokhorova, I, Yusupova, G, Yusupov, M. | | 登録日 | 2014-07-24 | | 公開日 | 2014-10-22 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (2.801 Å) | | 主引用文献 | Structural basis for the inhibition of the eukaryotic ribosome.
Nature, 513, 2014
|
|
4U4N
 
 | | Crystal structure of Edeine bound to the yeast 80S ribosome | | 分子名称: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | 著者 | Garreau de Loubresse, N, Prokhorova, I, Yusupova, G, Yusupov, M. | | 登録日 | 2014-07-24 | | 公開日 | 2014-10-22 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Structural basis for the inhibition of the eukaryotic ribosome.
Nature, 513, 2014
|
|
9J9I
 
 | |
