2HVQ
 
 | | Structure of Adenylated full-length T4 RNA Ligase 2 | | 分子名称: | Hypothetical 37.6 kDa protein in Gp24-hoc intergenic region, MAGNESIUM ION | | 著者 | Nandakumar, J, Lima, C.D. | | 登録日 | 2006-07-30 | | 公開日 | 2006-10-17 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | RNA Ligase Structures Reveal the Basis for RNA Specificity and Conformational Changes that Drive Ligation Forward.
Cell(Cambridge,Mass.), 127, 2006
|
|
1MG0
 
 | | Horse Liver Alcohol Dehydrogenase Complexed With NAD+ and 2,3-Difluorobenzyl Alcohol | | 分子名称: | 2,3-DIFLUOROBENZYL ALCOHOL, Alcohol Dehydrogenase E chain, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | 著者 | Rubach, J.K, Plapp, B.V. | | 登録日 | 2002-08-14 | | 公開日 | 2002-11-13 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Mobility of Fluorobenzyl Alcohols Bound to Liver Alcohol Dehydrogenases as Determined by NMR and X-ray Crystallographic Studies
Biochemistry, 41, 2002
|
|
1C9B
 
 | |
1NAM
 
 | | MURINE ALLOREACTIVE SCFV TCR-PEPTIDE-MHC CLASS I MOLECULE COMPLEX | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BM3.3 T Cell Receptor alpha-Chain, BM3.3 T Cell Receptor beta-Chain, ... | | 著者 | Reiser, J.-B, Darnault, C, Gregoire, C, Mosser, T, Mazza, G, Kearnay, A, van der Merwe, P.A, Fontecilla-Camps, J.C, Housset, D, Malissen, B. | | 登録日 | 2002-11-28 | | 公開日 | 2003-03-11 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | CDR3 loop flexibility contributes to the degeneracy of TCR recognition
Nat.Immunol., 4, 2003
|
|
1NJ4
 
 | |
2JV6
 
 | | YF ED3 Protein NMR Structure | | 分子名称: | Envelope protein E | | 著者 | Volk, D.E, Gandham, S.H.A, May, F.J, Anderson, A, Barrett, A.D.T, Gorenstein, D.G. | | 登録日 | 2007-09-12 | | 公開日 | 2008-09-16 | | 最終更新日 | 2022-03-16 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Yellow Fever Virus Envelope Protein Domain III: A Convergence of Structure and Phylogenetics
To be Published
|
|
1PTQ
 
 | | PROTEIN KINASE C DELTA CYS2 DOMAIN | | 分子名称: | PROTEIN KINASE C DELTA TYPE, ZINC ION | | 著者 | Zhang, G, Hurley, J.H. | | 登録日 | 1995-05-11 | | 公開日 | 1995-07-31 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Crystal structure of the cys2 activator-binding domain of protein kinase C delta in complex with phorbol ester.
Cell(Cambridge,Mass.), 81, 1995
|
|
2FH4
 
 | | C-terminal half of gelsolin soaked in EGTA at pH 8 | | 分子名称: | Gelsolin | | 著者 | Chumnarnsilpa, S, Loonchanta, A, Xue, B, Choe, H, Urosev, D, Wang, H, Burtnick, L.D, Robinson, R.C. | | 登録日 | 2005-12-23 | | 公開日 | 2006-06-13 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Calcium ion exchange in crystalline gelsolin
J.Mol.Biol., 357, 2006
|
|
1PTR
 
 | |
2FIK
 
 | | Structure of a microbial glycosphingolipid bound to mouse CD1d | | 分子名称: | (2S,3R)-3-HYDROXY-2-(TETRADECANOYLAMINO)OCTADECYL ALPHA-D-GALACTOPYRANOSIDURONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Wu, D, Zajonc, D.M. | | 登録日 | 2005-12-29 | | 公開日 | 2006-03-21 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Design of natural killer T cell activators: structure and function of a microbial glycosphingolipid bound to mouse CD1d.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2FK0
 
 | | Crystal Structure of a H5N1 influenza virus hemagglutinin. | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, hemagglutinin | | 著者 | Stevens, J, Wilson, I.A. | | 登録日 | 2006-01-03 | | 公開日 | 2006-05-02 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.95 Å) | | 主引用文献 | Structure and Receptor Specificity of the Hemagglutinin from an H5N1 Influenza Virus.
Science, 312, 2006
|
|
1AZC
 
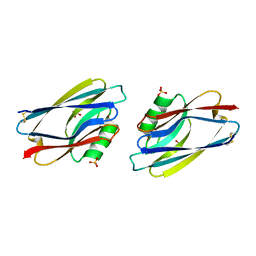 | | STRUCTURE OF APO-AZURIN FROM ALCALIGENES DENITRIFICANS AT 1.8 ANGSTROMS RESOLUTION | | 分子名称: | AZURIN, COPPER (II) ION, SULFATE ION | | 著者 | Baker, E.N, Shepard, W.E.B, Kingston, R.L. | | 登録日 | 1992-12-16 | | 公開日 | 1993-10-31 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure of apo-azurin from Alcaligenes denitrificans at 1.8 A resolution.
Acta Crystallogr.,Sect.D, 49, 1993
|
|
1PQ5
 
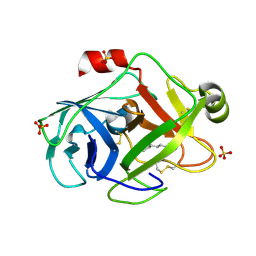 | | Trypsin at pH 5, 0.85 A | | 分子名称: | ARGININE, SULFATE ION, Trypsin | | 著者 | Schmidt, A, Jelsch, C, Rypniewski, W, Lamzin, V.S. | | 登録日 | 2003-06-18 | | 公開日 | 2003-11-11 | | 最終更新日 | 2017-10-11 | | 実験手法 | X-RAY DIFFRACTION (0.85 Å) | | 主引用文献 | Trypsin Revisited: CRYSTALLOGRAPHY AT (SUB) ATOMIC RESOLUTION AND QUANTUM CHEMISTRY REVEALING DETAILS OF CATALYSIS.
J.Biol.Chem., 278, 2003
|
|
1BRJ
 
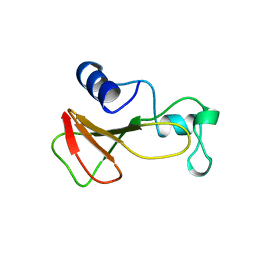 | | BARNASE MUTANT WITH ILE 88 REPLACED BY ALA | | 分子名称: | BARNASE, ZINC ION | | 著者 | Cramer, P.C, Buckle, A, Fersht, A. | | 登録日 | 1995-03-09 | | 公開日 | 1995-07-10 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural and energetic responses to cavity-creating mutations in hydrophobic cores: observation of a buried water molecule and the hydrophilic nature of such hydrophobic cavities.
Biochemistry, 35, 1996
|
|
1BRH
 
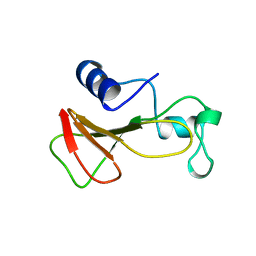 | | BARNASE MUTANT WITH LEU 14 REPLACED BY ALA | | 分子名称: | BARNASE, ZINC ION | | 著者 | Cramer, P.C, Buckle, A, Fersht, A. | | 登録日 | 1995-03-09 | | 公開日 | 1995-07-10 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural and energetic responses to cavity-creating mutations in hydrophobic cores: observation of a buried water molecule and the hydrophilic nature of such hydrophobic cavities.
Biochemistry, 35, 1996
|
|
1BSL
 
 | |
1Q45
 
 | | 12-0xo-phytodienoate reductase isoform 3 | | 分子名称: | 12-oxophytodienoate-10,11-reductase, FLAVIN MONONUCLEOTIDE | | 著者 | Phillips Jr, G.N, Johnson, K.A, Bingman, C.A, Smith, D.W, Center for Eukaryotic Structural Genomics (CESG) | | 登録日 | 2003-08-01 | | 公開日 | 2003-11-25 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | X-ray structure of Arabidopsis At2g06050, 12-oxophytodienoate reductase isoform 3
Proteins, 58, 2005
|
|
2JXY
 
 | | Solution structure of the hemopexin-like domain of MMP12 | | 分子名称: | CALCIUM ION, Macrophage metalloelastase | | 著者 | Bertini, I, Calderone, V, Fragai, M, Jaiswal, R, Luchinat, C, Melikian, M. | | 登録日 | 2007-12-01 | | 公開日 | 2008-05-27 | | 最終更新日 | 2022-03-16 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Evidence of reciprocal reorientation of the catalytic and hemopexin-like domains of full-length MMP-12
J.Am.Chem.Soc., 130, 2008
|
|
1PMC
 
 | |
1CE8
 
 | | CARBAMOYL PHOSPHATE SYNTHETASE FROM ESCHERICHIS COLI WITH COMPLEXED WITH THE ALLOSTERIC LIGAND IMP | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, INOSINIC ACID, ... | | 著者 | Thoden, J.B, Raushel, F.M, Holden, H.M. | | 登録日 | 1999-03-18 | | 公開日 | 1999-07-26 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | The binding of inosine monophosphate to Escherichia coli carbamoyl phosphate synthetase.
J.Biol.Chem., 274, 1999
|
|
2GUM
 
 | |
2HD5
 
 | | USP2 in complex with ubiquitin | | 分子名称: | Polyubiquitin, Ubiquitin carboxyl-terminal hydrolase 2, ZINC ION | | 著者 | Renatus, M, Kroemer, M. | | 登録日 | 2006-06-20 | | 公開日 | 2006-08-15 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structural Basis of Ubiquitin Recognition by the Deubiquitinating Protease USP2.
Structure, 14, 2006
|
|
2GL7
 
 | |
1N02
 
 | |
2GYR
 
 | | Crystal structure of human artemin | | 分子名称: | Neurotrophic factor artemin, isoform 3 | | 著者 | Wang, X.Q. | | 登録日 | 2006-05-09 | | 公開日 | 2006-06-27 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structure of Artemin Complexed with Its Receptor GFRalpha3: Convergent Recognition of Glial Cell Line-Derived Neurotrophic Factors.
Structure, 14, 2006
|
|
