1FBS
 
 | |
3SPA
 
 | | Crystal Structure of Human Mitochondrial RNA Polymerase | | 分子名称: | CHLORIDE ION, DNA-directed RNA polymerase, mitochondrial, ... | | 著者 | Ringel, R, Sologub, M, Morozov, Y.I, Litonin, D, Cramer, P, Temiakov, D. | | 登録日 | 2011-07-01 | | 公開日 | 2011-09-28 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structure of human mitochondrial RNA polymerase
Nature, 478, 2011
|
|
1AJY
 
 | | STRUCTURE AND MOBILITY OF THE PUT3 DIMER: A DNA PINCER, NMR, 13 STRUCTURES | | 分子名称: | PUT3, ZINC ION | | 著者 | Walters, K.J, Dayie, K.T, Reece, R.J, Ptashne, M, Wagner, G. | | 登録日 | 1997-05-12 | | 公開日 | 1997-09-17 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure and mobility of the PUT3 dimer.
Nat.Struct.Biol., 4, 1997
|
|
6E8E
 
 | |
6FVM
 
 | | Mutant DNA polymerase sliding clamp from Escherichia coli with bound P7 peptide | | 分子名称: | Beta sliding clamp, CALCIUM ION, GLYCEROL, ... | | 著者 | Martiel, I, Andre, C, Olieric, V, Guichard, G, Burnouf, D. | | 登録日 | 2018-03-04 | | 公開日 | 2019-04-10 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.631 Å) | | 主引用文献 | Peptide Interactions on Bacterial Sliding Clamps.
Acs Infect Dis., 2019
|
|
1S0V
 
 | | Structural basis for substrate selection by T7 RNA polymerase | | 分子名称: | 5'-D(*G*GP*GP*AP*AP*TP*CP*GP*AP*TP*AP*TP*CP*GP*CP*CP*GP*C)-3', 5'-D(*GP*TP*CP*GP*AP*TP*TP*CP*CP*C)-3', 5'-R(*AP*AP*CP*U*GP*CP*GP*GP*CP*GP*AP*U)-3', ... | | 著者 | Temiakov, D, Patlan, V, Anikin, M, McAllister, W.T, Yokoyama, S, Vassylyev, D.G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2004-01-05 | | 公開日 | 2004-02-24 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structural basis for substrate selection by t7 RNA polymerase.
Cell(Cambridge,Mass.), 116, 2004
|
|
1GKH
 
 | | MUTANT K69H OF GENE V PROTEIN (SINGLE-STRANDED DNA BINDING PROTEIN) | | 分子名称: | GENE V PROTEIN | | 著者 | Su, S, Gao, Y.-G, Zhang, H, Terwilliger, T.C, Wang, A.H.-J. | | 登録日 | 1997-03-04 | | 公開日 | 1997-09-04 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Analyses of the stability and function of three surface mutants (R82C, K69H, and L32R) of the gene V protein from Ff phage by X-ray crystallography.
Protein Sci., 6, 1997
|
|
5EIX
 
 | | QUINOLONE-STABILIZED CLEAVAGE COMPLEX OF TOPOISOMERASE IV FROM KLEBSIELLA PNEUMONIAE | | 分子名称: | (3S)-9-fluoro-3-methyl-10-(4-methylpiperazin-1-yl)-7-oxo-2,3-dihydro-7H-[1,4]oxazino[2,3,4-ij]quinoline-6-carboxylic acid, DNA topoisomerase 4 subunit B,DNA topoisomerase 4 subunit A, MAGNESIUM ION, ... | | 著者 | Veselkov, D.A, Laponogov, I, Pan, X.-S, Selvarajah, J, Branstrom, A, Fisher, L.M, Sanderson, M.R. | | 登録日 | 2015-10-30 | | 公開日 | 2016-04-13 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (3.35 Å) | | 主引用文献 | Structure of a quinolone-stabilized cleavage complex of topoisomerase IV from Klebsiella pneumoniae and comparison with a related Streptococcus pneumoniae complex.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
6BC8
 
 | | Crystal structure of Rev7-R124A/Rev3-RBM2 (residues 1988-2014) complex | | 分子名称: | ACETATE ION, DNA polymerase zeta catalytic subunit, Mitotic spindle assembly checkpoint protein MAD2B, ... | | 著者 | Rizzo, A.A, Hao, B, Li, Y, Korzhnev, D.M. | | 登録日 | 2017-10-20 | | 公開日 | 2018-08-01 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.68 Å) | | 主引用文献 | Rev7 dimerization is important for assembly and function of the Rev1/Pol zeta translesion synthesis complex.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
3ZKC
 
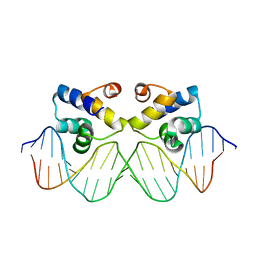 | | Crystal structure of the master regulator for biofilm formation SinR in complex with DNA. | | 分子名称: | 5'-D(*AP*AP*AP*GP*TP*TP*CP*TP*CP*TP*TP*TP*AP*GP *AP*GP*AP*AP*CP*AP*AP)-3', 5'-D(*AP*TP*TP*GP*TP*TP*CP*TP*CP*TP*AP*AP*AP*GP *AP*GP*AP*AP*CP*TP*TP)-3', HTH-TYPE TRANSCRIPTIONAL REGULATOR SINR | | 著者 | Newman, J.A, Rodrigues, C, Lewis, R.J. | | 登録日 | 2013-01-22 | | 公開日 | 2013-03-06 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Molecular Basis of the Activity of Sinr, the Master Regulator of Biofilm Formation in Bacillus Subtilis.
J.Biol.Chem., 288, 2013
|
|
6WS5
 
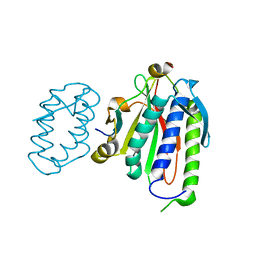 | | Rational drug design of phenazopyridine derivatives as novel inhibitors of Rev1-CT | | 分子名称: | 3-[(Z)-(2,3-difluorophenyl)diazenyl]pyridine-2,6-diamine, DNA polymerase zeta catalytic subunit, DNA repair protein REV1, ... | | 著者 | McPherson, K.S, Korzhnev, D.M. | | 登録日 | 2020-04-30 | | 公開日 | 2020-12-23 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.472 Å) | | 主引用文献 | Structure-Based Drug Design of Phenazopyridine Derivatives as Inhibitors of Rev1 Interactions in Translesion Synthesis.
Chemmedchem, 16, 2021
|
|
2O5I
 
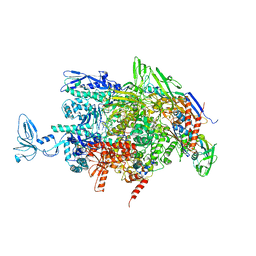 | | Crystal structure of the T. thermophilus RNA polymerase elongation complex | | 分子名称: | 5'-D(*AP*AP*CP*GP*CP*CP*AP*GP*AP*CP*AP*GP*GP*G)-3', 5'-D(P*CP*CP*CP*TP*GP*TP*CP*TP*GP*GP*CP*GP*TP*TP*CP*GP*CP*GP*CP*GP*CP*CP*G)-3', 5'-R(P*GP*AP*GP*UP*CP*UP*GP*CP*GP*GP*CP*GP*CP*GP*CP*G)-3', ... | | 著者 | Vassylyev, D.G, Tahirov, T.H, Vassylyeva, M.N. | | 登録日 | 2006-12-06 | | 公開日 | 2007-07-03 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural basis for transcription elongation by bacterial RNA polymerase.
Nature, 448, 2007
|
|
3P8B
 
 | |
6MKM
 
 | | Crystallographic solvent mapping analysis of DMSO/Tris bound to APE1 | | 分子名称: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DIMETHYL SULFOXIDE, ... | | 著者 | Georgiadis, M.M, He, H, Chen, Q. | | 登録日 | 2018-09-25 | | 公開日 | 2019-01-30 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.673 Å) | | 主引用文献 | Discovery of Macrocyclic Inhibitors of Apurinic/Apyrimidinic Endonuclease 1.
J. Med. Chem., 62, 2019
|
|
6MK3
 
 | | Crystallographic solvent mapping analysis of DMSO bound to APE1 | | 分子名称: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, DNA-(apurinic or apyrimidinic site) lyase | | 著者 | Georgiadis, M.M, He, H, Chen, Q. | | 登録日 | 2018-09-24 | | 公開日 | 2019-01-30 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.478 Å) | | 主引用文献 | Discovery of Macrocyclic Inhibitors of Apurinic/Apyrimidinic Endonuclease 1.
J. Med. Chem., 62, 2019
|
|
6MKK
 
 | | Crystallographic solvent mapping analysis of DMSO/Mg bound to APE1 | | 分子名称: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, DNA-(apurinic or apyrimidinic site) lyase, ... | | 著者 | Georgiadis, M.M, He, H, Chen, Q. | | 登録日 | 2018-09-25 | | 公開日 | 2019-01-30 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.442 Å) | | 主引用文献 | Discovery of Macrocyclic Inhibitors of Apurinic/Apyrimidinic Endonuclease 1.
J. Med. Chem., 62, 2019
|
|
1T2K
 
 | | Structure Of The DNA Binding Domains Of IRF3, ATF-2 and Jun Bound To DNA | | 分子名称: | 31-MER, Cyclic-AMP-dependent transcription factor ATF-2, Interferon regulatory factor 3, ... | | 著者 | Panne, D, Maniatis, T, Harrison, S.C. | | 登録日 | 2004-04-21 | | 公開日 | 2004-11-16 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Crystal structure of ATF-2/c-Jun and IRF-3 bound to the interferon-beta enhancer.
Embo J., 23, 2004
|
|
5YIU
 
 | |
1BW6
 
 | | HUMAN CENTROMERE PROTEIN B (CENP-B) DNA BINDIGN DOMAIN RP1 | | 分子名称: | PROTEIN (CENTROMERE PROTEIN B) | | 著者 | Iwahara, J, Kigawa, T, Kitagawa, K, Masumoto, H, Okazaki, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 1998-09-30 | | 公開日 | 1998-10-07 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | A helix-turn-helix structure unit in human centromere protein B (CENP-B).
EMBO J., 17, 1998
|
|
8OOS
 
 | | CryoEM Structure INO80core Hexasome complex ATPase-hexasome refinement state 2 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Chromatin-remodeling ATPase Ino80, DNA Strand 2, ... | | 著者 | Zhang, M, Jungblut, A, Hoffmann, T, Eustermann, S. | | 登録日 | 2023-04-05 | | 公開日 | 2023-07-26 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3.29 Å) | | 主引用文献 | Hexasome-INO80 complex reveals structural basis of noncanonical nucleosome remodeling.
Science, 381, 2023
|
|
4IPC
 
 | | Structure of the N-terminal domain of RPA70, E7R mutant | | 分子名称: | Replication protein A 70 kDa DNA-binding subunit | | 著者 | Feldkamp, M.D, Frank, A.O, Vangamudi, B, Fesik, S.W, Chazin, W.J. | | 登録日 | 2013-01-09 | | 公開日 | 2013-09-11 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.22 Å) | | 主引用文献 | Surface Reengineering of RPA70N Enables Cocrystallization with an Inhibitor of the Replication Protein A Interaction Motif of ATR Interacting Protein.
Biochemistry, 52, 2013
|
|
4IPD
 
 | | Structure of the N-terminal domain of RPA70, E100R mutant | | 分子名称: | Replication protein A 70 kDa DNA-binding subunit | | 著者 | Feldkamp, M.D, Frank, A.O, Vangamudi, B, Fesik, S.W, Chazin, W.J. | | 登録日 | 2013-01-09 | | 公開日 | 2013-09-11 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.51 Å) | | 主引用文献 | Surface Reengineering of RPA70N Enables Cocrystallization with an Inhibitor of the Replication Protein A Interaction Motif of ATR Interacting Protein.
Biochemistry, 52, 2013
|
|
4IPG
 
 | | Structure of the N-terminal domain of RPA70, E7R, E100R mutant | | 分子名称: | Replication protein A 70 kDa DNA-binding subunit | | 著者 | Feldkamp, M.D, Frank, A.O, Vangamudi, B, Fesik, S.W, Chazin, W.J. | | 登録日 | 2013-01-09 | | 公開日 | 2013-09-11 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.58 Å) | | 主引用文献 | Surface Reengineering of RPA70N Enables Cocrystallization with an Inhibitor of the Replication Protein A Interaction Motif of ATR Interacting Protein.
Biochemistry, 52, 2013
|
|
4IPH
 
 | | Structure of N-terminal domain of RPA70 in complex with VU079104 inhibitor | | 分子名称: | Replication protein A 70 kDa DNA-binding subunit, ~{N}-(2,3-dimethylphenyl)-7-oxidanylidene-12-sulfanylidene-5,11-dithia-1,8-diazatricyclo[7.3.0.0^{2,6}]dodeca-2(6),3,9-triene-10-carboxamide | | 著者 | Feldkamp, M.D, Frank, A.O, Vangamudi, B, Fesik, S.W, Chazin, W.J. | | 登録日 | 2013-01-09 | | 公開日 | 2013-09-11 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | Surface Reengineering of RPA70N Enables Cocrystallization with an Inhibitor of the Replication Protein A Interaction Motif of ATR Interacting Protein.
Biochemistry, 52, 2013
|
|
5E7N
 
 | | Crystal Structure of RPA70N in complex with VU0085636 | | 分子名称: | 2-({3-[(4-bromophenyl)sulfamoyl]-4-methylbenzoyl}amino)benzoic acid, Replication protein A 70 kDa DNA-binding subunit | | 著者 | Gilston, B.A, Patrone, J.D, Pelz, N.F, Bates, B.S, Souza-Fagundes, E.M, Vangamudi, B, Camper, D, Kuznetsov, A, Browning, C.F, Feldkamp, M.D, Olejniczak, E.T, Rossanese, O.W, Waterson, A.G, Fesik, S.W, Chazin, W.J. | | 登録日 | 2015-10-12 | | 公開日 | 2016-01-27 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.21 Å) | | 主引用文献 | Identification and Optimization of Anthranilic Acid Based Inhibitors of Replication Protein A.
Chemmedchem, 11, 2016
|
|
