4XID
 
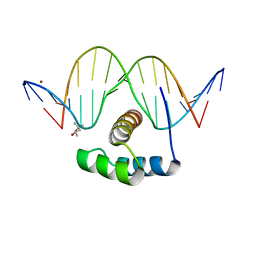 | | AntpHD with 15bp DNA duplex | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(*AP*GP*AP*AP*AP*GP*CP*CP*AP*TP*TP*AP*GP*AP*G)-3'), DNA (5'-D(*TP*CP*TP*CP*TP*AP*AP*TP*GP*GP*CP*TP*TP*TP*C)-3'), ... | | 著者 | White, M.A, Zandarashvili, L, Iwahara, J. | | 登録日 | 2015-01-06 | | 公開日 | 2015-11-25 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.701 Å) | | 主引用文献 | Entropic Enhancement of Protein-DNA Affinity by Oxygen-to-Sulfur Substitution in DNA Phosphate.
Biophys.J., 109, 2015
|
|
8PH4
 
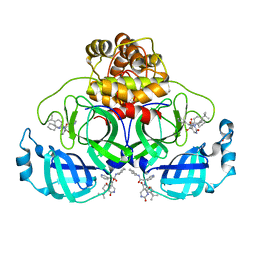 | | Co-Crystal structure of the SARS-CoV2 main protease Nsp5 with an Uracil-carrying X77-like inhibitor | | 分子名称: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, MALONATE ION, ... | | 著者 | Barthel, T, Altincekic, N, Jores, N, Wollenhaupt, J, Weiss, M.S, Schwalbe, H. | | 登録日 | 2023-06-18 | | 公開日 | 2024-01-31 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.69 Å) | | 主引用文献 | Targeting the Main Protease (M pro , nsp5) by Growth of Fragment Scaffolds Exploiting Structure-Based Methodologies.
Acs Chem.Biol., 19, 2024
|
|
6R5J
 
 | |
6L10
 
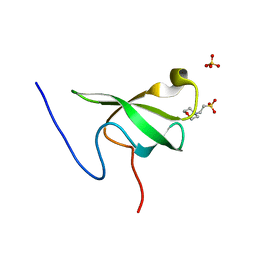 | | PHF20L1 Tudor1 - MES | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, PHD finger protein 20-like protein 1, SULFATE ION | | 著者 | Lv, M.Q, Gao, J. | | 登録日 | 2019-09-27 | | 公開日 | 2020-09-23 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Conformational Selection in Ligand Recognition by the First Tudor Domain of PHF20L1.
J Phys Chem Lett, 11, 2020
|
|
6L1F
 
 | |
6RM9
 
 | | Crystal structure of the DEAH-box ATPase Prp2 in complex with Spp2 and ADP | | 分子名称: | ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Hamann, F, Neumann, P, Ficner, R. | | 登録日 | 2019-05-06 | | 公開日 | 2020-02-05 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structural analysis of the intrinsically disordered splicing factor Spp2 and its binding to the DEAH-box ATPase Prp2.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6RMB
 
 | | Crystal structure of the DEAH-box ATPase Prp2 in complex with Spp2 and ADP | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Putative mRNA splicing factor, ... | | 著者 | Hamann, F, Neumann, P, Ficner, R. | | 登録日 | 2019-05-06 | | 公開日 | 2020-02-05 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural analysis of the intrinsically disordered splicing factor Spp2 and its binding to the DEAH-box ATPase Prp2.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5CRO
 
 | |
8IGD
 
 | |
8PHL
 
 | | Human carbonic anhydrase II containing 4-fluorophenylalanine | | 分子名称: | Carbonic anhydrase 2, MERCURIBENZOIC ACID, ZINC ION | | 著者 | Pham, L.B.T, Costantino, A, Barbieri, L, Calderone, V, Luchinat, E, Banci, L. | | 登録日 | 2023-06-20 | | 公開日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Controlling the incorporation of fluorinated amino acids in human cells and its structural impact.
Protein Sci., 33, 2024
|
|
3T7C
 
 | |
5GHL
 
 | | Crystal structure Analysis of the starch-binding domain of glucoamylase from Aspergillus niger | | 分子名称: | GLYCEROL, Glucoamylase, SULFATE ION | | 著者 | Miyake, H, Suyama, Y, Muraki, N, Kusunoki, M, Tanaka, A. | | 登録日 | 2016-06-20 | | 公開日 | 2017-10-18 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of the starch-binding domain of glucoamylase from Aspergillus niger.
Acta Crystallogr.,Sect.F, 73, 2017
|
|
5EJ2
 
 | |
3S55
 
 | |
3TSC
 
 | |
3SX2
 
 | |
4V5L
 
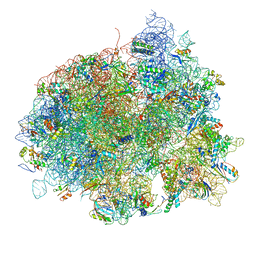 | | The structure of EF-Tu and aminoacyl-tRNA bound to the 70S ribosome with a GTP analog | | 分子名称: | 16S RRNA, 23S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | 著者 | Voorhees, R.M, Schmeing, T.M, Ramakrishnan, V. | | 登録日 | 2010-09-02 | | 公開日 | 2014-07-09 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | The Mechanism for Activation of GTP Hydrolysis on the Ribosome.
Science, 330, 2010
|
|
6DKQ
 
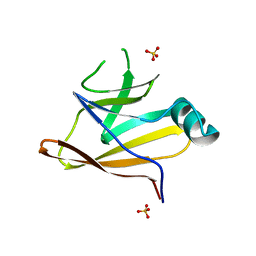 | | Crystal structure of the Shr Hemoglobin Interacting Domain 2 | | 分子名称: | Heme-binding protein Shr, SULFATE ION | | 著者 | Macdonald, R, Cascio, D, Collazo, M.J, Clubb, R.T. | | 登録日 | 2018-05-30 | | 公開日 | 2018-10-24 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | The Streptococcus pyogenes Shr protein captures human hemoglobin using two structurally unique binding domains.
J.Biol.Chem., 293, 2018
|
|
6DZ8
 
 | |
6EGS
 
 | | Crystal structure of the GalNAc-T2 F104S mutant in complex with UDP-GalNAc | | 分子名称: | MANGANESE (II) ION, Polypeptide N-acetylgalactosaminyltransferase 2, URIDINE-DIPHOSPHATE-N-ACETYLGALACTOSAMINE | | 著者 | de las Rivas, M, Coelho, H, Diniz, A, Lira-Navarrete, E, Jimenez-Barbero, J, Schjoldager, K.T, Bennett, E.P, Vakhrushev, S.Y, Clausen, H, Corzana, F, Marcelo, F, Hurtado-Guerrero, R. | | 登録日 | 2017-09-12 | | 公開日 | 2018-04-11 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structural Analysis of a GalNAc-T2 Mutant Reveals an Induced-Fit Catalytic Mechanism for GalNAc-Ts.
Chemistry, 24, 2018
|
|
6EJX
 
 | |
3OEC
 
 | |
3PGX
 
 | |
3PXX
 
 | |
4KZ3
 
 | | Crystal structure of AmpC beta-lactamase in complex with fragment 44 (5-chloro-3-sulfamoylthiophene-2-carboxylic acid) | | 分子名称: | 5-chloro-3-sulfamoylthiophene-2-carboxylic acid, Beta-lactamase, PHOSPHATE ION | | 著者 | Eidam, O, Barelier, S, Fish, I, Shoichet, B.K. | | 登録日 | 2013-05-29 | | 公開日 | 2014-05-21 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.67 Å) | | 主引用文献 | Increasing chemical space coverage by combining empirical and computational fragment screens.
Acs Chem.Biol., 9, 2014
|
|
