4YAX
 
 | | Crystal structure of TRIM24 PHD-bromodomain complexed with N-[6-(4-methoxyphenoxy)-1,3-dimethyl-2-oxo-2,3-dihydro-1H-1,3-benzodiazol-5-yl]benzenesulfonamide (5g) | | 分子名称: | GLYCEROL, N-[6-(4-methoxyphenoxy)-1,3-dimethyl-2-oxo-2,3-dihydro-1H-benzimidazol-5-yl]benzenesulfonamide, SULFATE ION, ... | | 著者 | Poncet-Montange, G, Palmer, W, Jones, P. | | 登録日 | 2015-02-18 | | 公開日 | 2015-06-24 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Structure-Guided Design of IACS-9571, a Selective High-Affinity Dual TRIM24-BRPF1 Bromodomain Inhibitor.
J.Med.Chem., 59, 2016
|
|
5R87
 
 | |
5R8L
 
 | |
4YAT
 
 | | Crystal structure of TRIM24 PHD-bromodomain complexed with N-(1,3-dimethyl-2-oxo-2,3-dihydro-1H-1,3-benzodiazol-5-yl)-4-methoxybenzene-1-sulfonamide (5b) | | 分子名称: | N-(1,3-dimethyl-2-oxo-2,3-dihydro-1H-benzimidazol-5-yl)-4-methoxybenzenesulfonamide, Transcription intermediary factor 1-alpha, ZINC ION | | 著者 | Poncet-Montange, G, Palmer, W, Jones, P. | | 登録日 | 2015-02-17 | | 公開日 | 2015-06-24 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.18 Å) | | 主引用文献 | Structure-Guided Design of IACS-9571, a Selective High-Affinity Dual TRIM24-BRPF1 Bromodomain Inhibitor.
J.Med.Chem., 59, 2016
|
|
4YBT
 
 | | Crystal structure of TRIM24 PHD-bromodomain complexed with N-{1,3-dimethyl-2-oxo-6-[3-(oxolan-3-ylmethoxy)phenoxy]-2,3-dihydro-1H-1,3-benzodiazol-5-yl}-1-methyl-1H-imidazole-4-sulfonamide (7l) | | 分子名称: | DIMETHYL SULFOXIDE, N-(1,3-dimethyl-2-oxo-6-{3-[(3S)-tetrahydrofuran-3-ylmethoxy]phenoxy}-2,3-dihydro-1H-benzimidazol-5-yl)-1-methyl-1H-imidazole-4-sulfonamide, Transcription intermediary factor 1-alpha, ... | | 著者 | Poncet-Montange, G, Palmer, W, Jones, P. | | 登録日 | 2015-02-19 | | 公開日 | 2015-06-24 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.82 Å) | | 主引用文献 | Structure-Guided Design of IACS-9571, a Selective High-Affinity Dual TRIM24-BRPF1 Bromodomain Inhibitor.
J.Med.Chem., 59, 2016
|
|
4YC9
 
 | | Crystal structure of TRIM24 PHD-bromodomain complexed with N-(6-{3-[4-(dimethylamino)butoxy]-5-propoxyphenoxy}-1,3-dimethyl-2-oxo-2,3-dihydro-1H-1,3-benzodiazol-5-yl)-3,4-dimethoxybenzene-1-sulfonamide (8i) | | 分子名称: | GLYCEROL, N-(6-{3-[4-(dimethylamino)butoxy]-5-propoxyphenoxy}-1,3-dimethyl-2-oxo-2,3-dihydro-1H-benzimidazol-5-yl)-3,4-dimethoxybenzenesulfonamide, Transcription intermediary factor 1-alpha, ... | | 著者 | Poncet-Montange, G, Palmer, W, Jones, P. | | 登録日 | 2015-02-19 | | 公開日 | 2015-06-24 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.82 Å) | | 主引用文献 | Structure-Guided Design of IACS-9571, a Selective High-Affinity Dual TRIM24-BRPF1 Bromodomain Inhibitor.
J.Med.Chem., 59, 2016
|
|
7T40
 
 | | Structure of MERS 3CL protease in complex with inhibitor 10c | | 分子名称: | (1R,2S)-1-hydroxy-2-{[N-({[7-(methanesulfonyl)-7-azaspiro[3.5]nonan-2-yl]oxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-1-hydroxy-2-{[N-({[7-(methanesulfonyl)-7-azaspiro[3.5]nonan-2-yl]oxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase, ... | | 著者 | Lovell, S, Liu, L, Battaile, K.P, Chamandi, S.D, Kim, Y, Groutas, W.C, Chang, K.O. | | 登録日 | 2021-12-09 | | 公開日 | 2021-12-22 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structure-Guided Design of Potent Spirocyclic Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3C-like Protease.
J.Med.Chem., 65, 2022
|
|
7T3Z
 
 | | Structure of MERS 3CL protease in complex with inhibitor 9c | | 分子名称: | (1R,2S)-2-{[N-({[(2r,4R)-7-acetyl-7-azaspiro[3.5]non-5-en-2-yl]oxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-2-{[N-({[7-(phenylacetyl)-7-azaspiro[3.5]nonan-2-yl]oxy}carbonyl)-L-leucyl]amino}propane-1-sulfonic acid, 3C-like proteinase | | 著者 | Lovell, S, Liu, L, Battaile, K.P, Chamandi, S.D, Kim, Y, Groutas, W.C, Chang, K.O. | | 登録日 | 2021-12-09 | | 公開日 | 2021-12-22 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structure-Guided Design of Potent Spirocyclic Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3C-like Protease.
J.Med.Chem., 65, 2022
|
|
7T3Y
 
 | | Structure of MERS 3CL protease in complex with inhibitor 8c | | 分子名称: | (1R,2S)-1-hydroxy-2-{[N-({[7-(2-methylpropanoyl)-7-azaspiro[3.5]nonan-2-yl]oxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-1-hydroxy-2-{[N-({[7-(2-methylpropanoyl)-7-azaspiro[3.5]nonan-2-yl]oxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase | | 著者 | Lovell, S, Liu, L, Battaile, K.P, Chamandi, S.D, Kim, Y, Groutas, W.C, Chang, K.O. | | 登録日 | 2021-12-09 | | 公開日 | 2021-12-22 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure-Guided Design of Potent Spirocyclic Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3C-like Protease.
J.Med.Chem., 65, 2022
|
|
7T41
 
 | | Structure of MERS 3CL protease in complex with inhibitor 14c | | 分子名称: | (1R,2S)-2-{[N-({[1-(tert-butoxycarbonyl)azetidin-3-yl]methoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3R)-2-oxo-2,3-dihydro-1H-pyrrol-3-yl]propane-1-sulfonic acid, (1S,2S)-2-{[N-({[1-(tert-butoxycarbonyl)azetidin-3-yl]methoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3R)-2-oxo-2,3-dihydro-1H-pyrrol-3-yl]propane-1-sulfonic acid, 3C-like proteinase | | 著者 | Lovell, S, Kashipathy, M.M, Battaile, K.P, Chamandi, S.D, Kim, Y, Groutas, W.C, Chang, K.O. | | 登録日 | 2021-12-09 | | 公開日 | 2021-12-22 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure-Guided Design of Potent Spirocyclic Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3C-like Protease.
J.Med.Chem., 65, 2022
|
|
7B9Y
 
 | | Structure of the FKBP51FK1 domain in complex with the macrocyclic SAFit analogue 64a | | 分子名称: | 2-cyclohexyl-12-[2-(3,4-dimethoxyphenyl)ethyl]-20,21-dihydroxy-25,26-dimethoxy-11,18,23-trioxa-4-azatetracyclo[22.3.1.113,17.04,9]nonacosa-1(27),13(29),14,16,24(28),25-hexaene-3,10-dione, Peptidyl-prolyl cis-trans isomerase FKBP5 | | 著者 | Bauder, M, Meyners, C, Purder, P, Merz, S, Voll, A, Heymann, T, Hausch, F. | | 登録日 | 2020-12-15 | | 公開日 | 2021-03-17 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Structure-Based Design of High-Affinity Macrocyclic FKBP51 Inhibitors.
J.Med.Chem., 64, 2021
|
|
7BA0
 
 | | Structure of the FKBP51FK1 domain in complex with the macrocyclic SAFit analogue 63 | | 分子名称: | 2-cyclohexyl-12-[2-(3,4-dimethoxyphenyl)ethyl]-20,21-dihydroxy-25,28-dimethoxy-11,18,23-trioxa-4-azatetracyclo[22.2.2.113,17.04,9]nonacosa-1(26),13(29),14,16,24,27-hexaene-3,10-dione, Peptidyl-prolyl cis-trans isomerase FKBP5 | | 著者 | Bauder, M, Meyners, C, Purder, P, Merz, S, Voll, A, Heymann, T, Hausch, F. | | 登録日 | 2020-12-15 | | 公開日 | 2021-03-17 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.14 Å) | | 主引用文献 | Structure-Based Design of High-Affinity Macrocyclic FKBP51 Inhibitors.
J.Med.Chem., 64, 2021
|
|
7B9Z
 
 | | Structure of the FKBP51FK1 domain in complex with the macrocyclic SAFit analogue 35-(E) | | 分子名称: | 2-cyclohexyl-12-[2-(3,4-dimethoxyphenyl)ethyl]-25,26-dimethoxy-11,18,23-trioxa-4-azatetracyclo[22.3.1.113,17.04,9]nonacosa-1(27),13(29),14,16,20,24(28),25-heptaene-3,10-dione, Peptidyl-prolyl cis-trans isomerase FKBP5, isothiocyanate | | 著者 | Bauder, M, Meyners, C, Purder, P, Merz, S, Voll, A, Heymann, T, Hausch, F. | | 登録日 | 2020-12-15 | | 公開日 | 2021-03-17 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.44 Å) | | 主引用文献 | Structure-Based Design of High-Affinity Macrocyclic FKBP51 Inhibitors.
J.Med.Chem., 64, 2021
|
|
7K7Q
 
 | |
7K7O
 
 | |
5R88
 
 | |
5R8P
 
 | |
5R8J
 
 | |
5R8F
 
 | |
5R86
 
 | |
5R8H
 
 | |
5R8K
 
 | |
7KQD
 
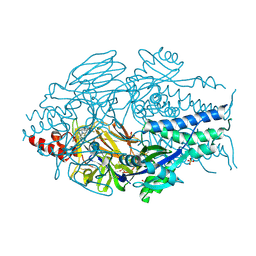 | | Prefusion RSV F Bound to RV521 | | 分子名称: | 1'-{[5-(aminomethyl)-1-(4,4,4-trifluorobutyl)-1H-benzimidazol-2-yl]methyl}-6'-fluorospiro[cyclopropane-1,3'-indol]-2'(1'H)-one, Fusion glycoprotein F0, SULFATE ION | | 著者 | McLellan, J.S. | | 登録日 | 2020-11-14 | | 公開日 | 2021-04-21 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.94 Å) | | 主引用文献 | Discovery of Sisunatovir (RV521), an Inhibitor of Respiratory Syncytial Virus Fusion.
J.Med.Chem., 64, 2021
|
|
5R85
 
 | |
5R8G
 
 | |
