4WFM
 
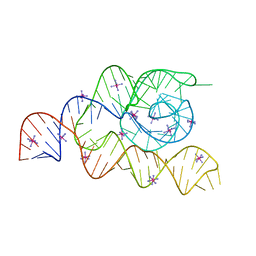 | | Structure of the complete bacterial SRP Alu domain | | 分子名称: | Bacillus subtilis small cytoplasmic RNA (scRNA),RNA, COBALT HEXAMMINE(III), MAGNESIUM ION | | 著者 | Kempf, G, Wild, K, Sinning, I. | | 登録日 | 2014-09-15 | | 公開日 | 2014-10-15 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Structure of the complete bacterial SRP Alu domain.
Nucleic Acids Res., 42, 2014
|
|
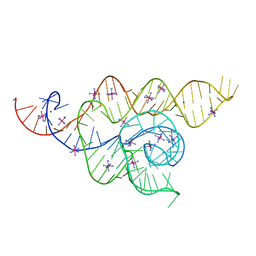
4WFL
 
 | |
1JSA
 
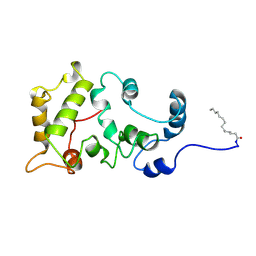 | | MYRISTOYLATED RECOVERIN WITH TWO CALCIUMS BOUND, NMR, 24 STRUCTURES | | 分子名称: | CALCIUM ION, MYRISTIC ACID, RECOVERIN | | 著者 | Ames, J.B, Ishima, R, Tanaka, T, Gordon, J.I, Stryer, L, Ikura, M. | | 登録日 | 1997-06-04 | | 公開日 | 1997-10-15 | | 最終更新日 | 2022-02-23 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Molecular mechanics of calcium-myristoyl switches.
Nature, 389, 1997
|
|
1QQC
 
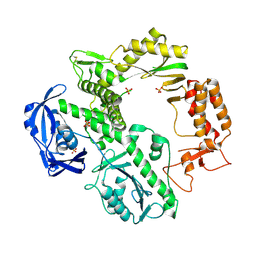 | | CRYSTAL STRUCTURE OF AN ARCHAEBACTERIAL DNA POLYMERASE D.TOK | | 分子名称: | DNA POLYMERASE II, MAGNESIUM ION, SULFATE ION | | 著者 | Zhao, Y, Jeruzalmi, D, Leighton, L, Lasken, R, Kuriyan, J. | | 登録日 | 1999-06-02 | | 公開日 | 1999-10-14 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Crystal structure of an archaebacterial DNA polymerase
Structure Fold.Des., 7, 1999
|
|
7T81
 
 | |
7T7C
 
 | |
7T7R
 
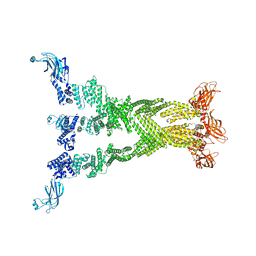 | |
7T7X
 
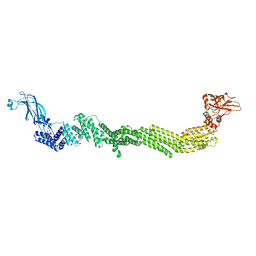 | |
7T7V
 
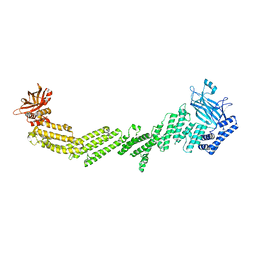 | |
5SUQ
 
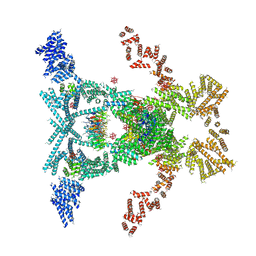 | | Crystal structure of the THO-Sub2 complex | | 分子名称: | 12-TUNGSTOPHOSPHATE, ATP-dependent RNA helicase SUB2, Tex1, ... | | 著者 | Ren, Y, Blobel, G. | | 登録日 | 2016-08-03 | | 公開日 | 2017-01-18 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (6 Å) | | 主引用文献 | Structural and biochemical analyses of the DEAD-box ATPase Sub2 in association with THO or Yra1.
Elife, 6, 2017
|
|
5SUP
 
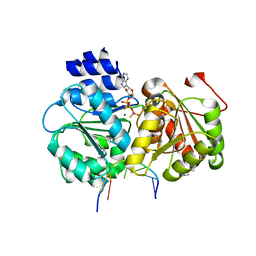 | | Crystal structure of the Sub2-Yra1 complex in association with RNA | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent RNA helicase SUB2, BERYLLIUM TRIFLUORIDE ION, ... | | 著者 | Ren, Y, Schmiege, P, Blobel, G. | | 登録日 | 2016-08-03 | | 公開日 | 2017-01-18 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural and biochemical analyses of the DEAD-box ATPase Sub2 in association with THO or Yra1.
Elife, 6, 2017
|
|
2LPA
 
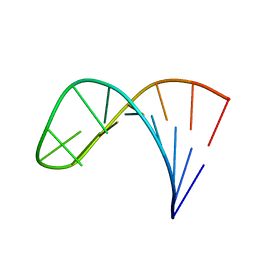 | | Mutant of the sub-genomic promoter from Brome Mosaic Virus | | 分子名称: | RNA (5'-R(*GP*AP*GP*GP*AP*CP*AP*UP*AP*GP*UP*CP*UP*UP*C)-3') | | 著者 | Skov, J. | | 登録日 | 2012-02-06 | | 公開日 | 2012-05-16 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The subgenomic promoter of brome mosaic virus folds into a stem-loop structure capped by a pseudo-triloop that is structurally similar to the triloop of the genomic promoter.
Rna, 18, 2012
|
|
2LP9
 
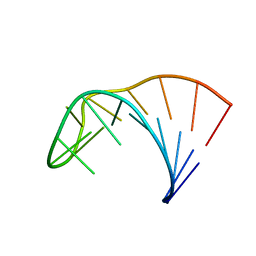 | | Pseudo-triloop from the sub-genomic promoter of Brome Mosaic Virus | | 分子名称: | RNA (5'-R(*GP*AP*GP*GP*AP*CP*AP*UP*AP*GP*AP*UP*CP*UP*UP*C)-3') | | 著者 | Skov, J. | | 登録日 | 2012-02-06 | | 公開日 | 2012-05-16 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The subgenomic promoter of brome mosaic virus folds into a stem-loop structure capped by a pseudo-triloop that is structurally similar to the triloop of the genomic promoter.
Rna, 18, 2012
|
|
7UJB
 
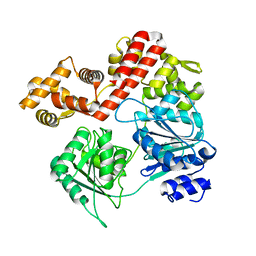 | | N-terminal domain deletion variant of Eta | | 分子名称: | DEAD/DEAH box RNA helicase | | 著者 | Qayyum, M.Z, Murakami, K.S. | | 登録日 | 2022-03-30 | | 公開日 | 2022-08-10 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (4.12 Å) | | 主引用文献 | The structure and activities of the archaeal transcription termination factor Eta detail vulnerabilities of the transcription elongation complex.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7TPT
 
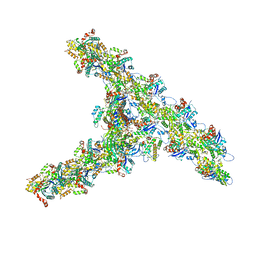 | | Single-particle Cryo-EM structure of Arp2/3 complex at branched-actin junction. | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | 著者 | Ding, B, Narvaez-Ortiz, H.Y, Nolen, B.J, Chowdhury, S. | | 登録日 | 2022-01-26 | | 公開日 | 2022-05-25 | | 最終更新日 | 2022-06-08 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Structure of Arp2/3 complex at a branched actin filament junction resolved by single-particle cryo-electron microscopy.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6XRA
 
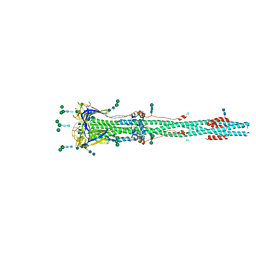 | | Distinct conformational states of SARS-CoV-2 spike protein | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | 著者 | Zhang, J, Cai, Y.F, Xiao, T.S, Peng, H.Q, Sterling, S.M, Walsh Jr, R.M, Rawson, S, Rits-Volloch, S, Chen, B. | | 登録日 | 2020-07-11 | | 公開日 | 2020-07-22 | | 最終更新日 | 2020-10-07 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Distinct conformational states of SARS-CoV-2 spike protein.
Science, 369, 2020
|
|
6XR8
 
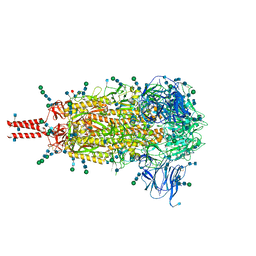 | | Distinct conformational states of SARS-CoV-2 spike protein | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Zhang, J, Cai, Y.F, Xiao, T.S, Peng, H.Q, Sterling, S.M, Walsh Jr, R.M, Rawson, S, Volloch, S.R, Chen, B. | | 登録日 | 2020-07-11 | | 公開日 | 2020-07-22 | | 最終更新日 | 2020-11-25 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Distinct conformational states of SARS-CoV-2 spike protein.
Science, 369, 2020
|
|
8TOE
 
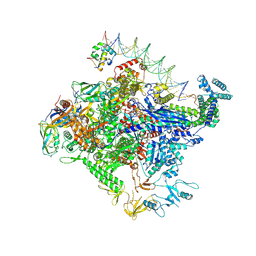 | |
8TO6
 
 | | Escherichia coli RNA polymerase unwinding intermediate (I1d) at the lambda PR promoter | | 分子名称: | (3R,5S,7R,8R,9S,10S,12S,13R,14S,17R)-10,13-dimethyl-17-[(2R)-pentan-2-yl]-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthrene-3,7,12-triol, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | 著者 | Darst, S.A, Saecker, R.M, Mueller, A.U. | | 登録日 | 2023-08-02 | | 公開日 | 2024-07-03 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Early intermediates in bacterial RNA polymerase promoter melting visualized by time-resolved cryo-electron microscopy.
Nat.Struct.Mol.Biol., 2024
|
|
8TOM
 
 | |
8TO8
 
 | | Escherichia coli RNA polymerase unwinding intermediate (I1b) at the lambda PR promoter | | 分子名称: | (3R,5S,7R,8R,9S,10S,12S,13R,14S,17R)-10,13-dimethyl-17-[(2R)-pentan-2-yl]-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthrene-3,7,12-triol, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | 著者 | Darst, S.A, Saecker, R.M, Mueller, A.U. | | 登録日 | 2023-08-03 | | 公開日 | 2024-07-03 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Early intermediates in bacterial RNA polymerase promoter melting visualized by time-resolved cryo-electron microscopy.
Nat.Struct.Mol.Biol., 2024
|
|
8OPP
 
 | | Structure of human terminal uridylyltransferase 7 (hTUT7/ZCCHC6) bound with pre-let7g miRNA and UTPalphaS | | 分子名称: | RNA (25-MER), Terminal uridylyltransferase 7, [[(2~{R},3~{S},4~{R},5~{R})-5-[2,4-bis(oxidanylidene)pyrimidin-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-sulfanyl-phosphoryl] phosphono hydrogen phosphate | | 著者 | Yi, G, Ye, M, Gilbert, R.J. | | 登録日 | 2023-04-07 | | 公開日 | 2024-07-24 | | 最終更新日 | 2024-08-07 | | 実験手法 | ELECTRON MICROSCOPY (3.76 Å) | | 主引用文献 | Structural basis for activity switching in polymerases determining the fate of let-7 pre-miRNAs.
Nat.Struct.Mol.Biol., 2024
|
|
8OPS
 
 | |
8OST
 
 | | Structure of human terminal uridylyltransferase 4 (TUT4, ZCCHC11) in complex with pre-let7g miRNA and Lin28A | | 分子名称: | Protein lin-28 homolog A, Terminal uridylyltransferase 4, ZINC ION, ... | | 著者 | Gilbert, R.J, Yi, G, Ye, M. | | 登録日 | 2023-04-20 | | 公開日 | 2024-07-17 | | 最終更新日 | 2024-08-07 | | 実験手法 | ELECTRON MICROSCOPY (3.69 Å) | | 主引用文献 | Structural basis for activity switching in polymerases determining the fate of let-7 pre-miRNAs.
Nat.Struct.Mol.Biol., 2024
|
|
8OPT
 
 | |
