2VSF
 
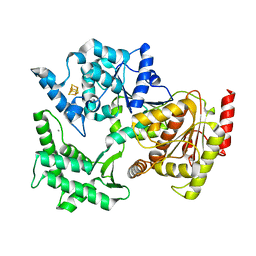 | | Structure of XPD from Thermoplasma acidophilum | | 分子名称: | CALCIUM ION, DNA REPAIR HELICASE RAD3 RELATED PROTEIN, IRON/SULFUR CLUSTER | | 著者 | Kuper, J, Wolski, S.C, Truglio, J.J, Kisker, C. | | 登録日 | 2008-04-23 | | 公開日 | 2008-07-08 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Crystal Structure of the Fes Cluster-Containing Nucleotide Excision Repair Helicase Xpd.
Plos Biol., 6, 2008
|
|
7VZ4
 
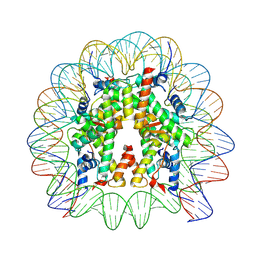 | | Cryo-EM structure of human nucleosome core particle composed of the Widom 601L DNA sequence | | 分子名称: | DNA (145-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | 著者 | Takizawa, Y, Ho, C.-H, Sato, S, Danev, R, Kurumizaka, H. | | 登録日 | 2021-11-15 | | 公開日 | 2023-05-17 | | 最終更新日 | 2024-06-26 | | 実験手法 | ELECTRON MICROSCOPY (1.89 Å) | | 主引用文献 | Methods for High Resolution Cryo-EM Analyses of Nucleosomes
To Be Published
|
|
7DO1
 
 | |
2B2U
 
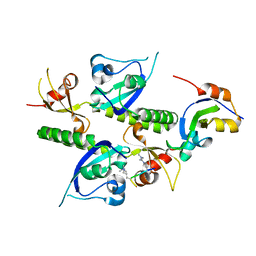 | | Tandem chromodomains of human CHD1 complexed with Histone H3 Tail containing trimethyllysine 4 and dimethylarginine 2 | | 分子名称: | Chromodomain-helicase-DNA-binding protein 1, Histone H3 | | 著者 | Flanagan IV, J.F, Mi, L.-Z, Chruszcz, M, Cymborowski, M, Clines, K.L, Kim, Y, Minor, W, Rastinejad, F, Khorasanizadeh, S. | | 登録日 | 2005-09-19 | | 公開日 | 2005-12-27 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.95 Å) | | 主引用文献 | Double chromodomains cooperate to recognize the methylated histone H3 tail.
Nature, 438, 2005
|
|
8BV6
 
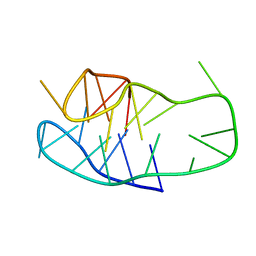 | | An i-motif domain able to undergo pH-dependent conformational transitions (neutral structure) | | 分子名称: | DNA (5'-D(*CP*(DNR)P*GP*TP*TP*CP*(DNR)P*GP*TP*TP*TP*TP*TP*CP*CP*GP*TP*TP*CP*CP*GP*T)-3') | | 著者 | Serrano-Chacon, I, Mir, B, Cupellini, L, Colizzi, F, Orozco, M, Escaja, N, Gonzalez, C. | | 登録日 | 2022-12-01 | | 公開日 | 2023-02-22 | | 最終更新日 | 2024-06-19 | | 実験手法 | SOLUTION NMR | | 主引用文献 | pH-Dependent Capping Interactions Induce Large-Scale Structural Transitions in i-Motifs.
J.Am.Chem.Soc., 145, 2023
|
|
2B2V
 
 | | Crystal structure analysis of human CHD1 chromodomains 1 and 2 bound to histone H3 resi 1-15 MeK4 | | 分子名称: | Chromodomain-helicase-DNA-binding protein 1, Histone H3 | | 著者 | Flanagan IV, J.F, Mi, L.-Z, Chruszcz, M, Cymborowski, M, Clines, K.L, Kim, Y, Minor, W, Rastinejad, F, Khorasanizadeh, S. | | 登録日 | 2005-09-19 | | 公開日 | 2005-12-27 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Double chromodomains cooperate to recognize the methylated histone H3 tail.
Nature, 438, 2005
|
|
2B2Y
 
 | | Tandem chromodomains of human CHD1 | | 分子名称: | Chromodomain-helicase-DNA-binding protein 1 | | 著者 | Flanagan IV, J.F, Mi, L.-Z, Chruszcz, M, Cymborowski, M, Clines, K.L, Kim, Y, Minor, W, Rastinejad, F, Khorasanizadeh, S. | | 登録日 | 2005-09-19 | | 公開日 | 2005-12-27 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Double chromodomains cooperate to recognize the methylated histone H3 tail.
Nature, 438, 2005
|
|
6KN4
 
 | |
6UPH
 
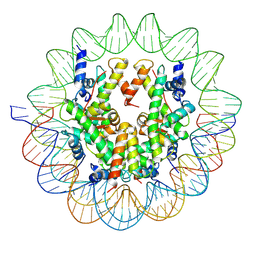 | | Structure of a Yeast Centromeric Nucleosome at 2.7 Angstrom resolution | | 分子名称: | DNA (119-MER), Histone H2A, Histone H2B.1, ... | | 著者 | Migl, D, Kschonsak, M, Arthur, C.P, Khin, Y, Harrison, S.C, Ciferri, C, Dimitrova, Y.N. | | 登録日 | 2019-10-17 | | 公開日 | 2019-11-06 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Cryoelectron Microscopy Structure of a Yeast Centromeric Nucleosome at 2.7 angstrom Resolution.
Structure, 28, 2020
|
|
8F5C
 
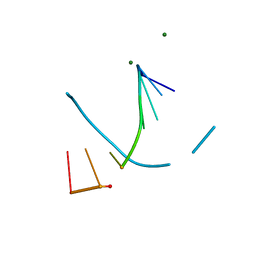 | | Mirror-image DNA containing 2'-OMe-L-dC modification | | 分子名称: | DNA (5'-D(*(0DG))-R(P*(XE6))-D(P*(0DG)P*(0DT)P*(0DA)P*(0DC)P*(0DG)P*(0DC))-3'), MAGNESIUM ION | | 著者 | Zhang, W, Dantsu, Y. | | 登録日 | 2022-11-13 | | 公開日 | 2023-09-20 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.15 Å) | | 主引用文献 | Synthesis and Structural Characterization of 2'-Deoxy-2'-Methoxy-L-Cytidine Nucleic Acids
Chemistryselect, 2023
|
|
3WVH
 
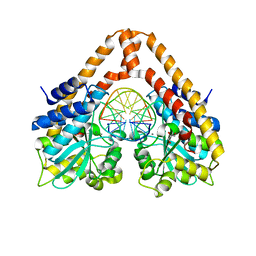 | | Time-Resolved Crystal Structure of HindIII with 25sec soaking | | 分子名称: | DNA (5'-D(*GP*CP*CP*AP*AP*GP*CP*TP*TP*GP*GP*C)-3'), GLYCEROL, MANGANESE (II) ION, ... | | 著者 | Kawamura, T, Kobayashi, T, Watanabe, N. | | 登録日 | 2014-05-21 | | 公開日 | 2015-04-08 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.54 Å) | | 主引用文献 | Analysis of the HindIII-catalyzed reaction by time-resolved crystallography
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6PNK
 
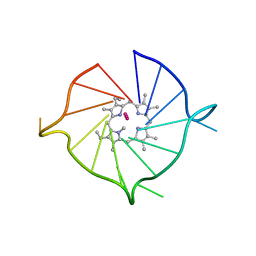 | |
3WVI
 
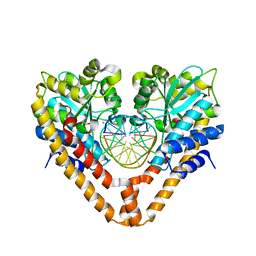 | | Time-Resolved Crystal Structure of HindIII with 40 sec soaking | | 分子名称: | DNA (5'-D(*GP*CP*CP*AP*AP*GP*CP*TP*TP*GP*GP*C)-3'), GLYCEROL, MANGANESE (II) ION, ... | | 著者 | Kawamura, T, Kobayashi, T, Watanabe, N. | | 登録日 | 2014-05-21 | | 公開日 | 2015-04-08 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Analysis of the HindIII-catalyzed reaction by time-resolved crystallography
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6FC9
 
 | | The 1,8-bis(aminomethyl)anthracene and Quadruplex-duplex junction complex | | 分子名称: | DNA (27-MER), [8-(azaniumylmethyl)anthracen-1-yl]methylazanium | | 著者 | Santana, A, Serrano, I, Montalvillo-Jimenez, L, Corzana, F, Bastida, A, Jimenez-Barbero, J, Gonzalez, C, Asensio, J.L. | | 登録日 | 2017-12-20 | | 公開日 | 2019-04-10 | | 最終更新日 | 2024-06-19 | | 実験手法 | SOLUTION NMR | | 主引用文献 | De Novo Design of Selective Quadruplex-Duplex Junction Ligands and Structural Characterisation of Their Binding Mode: Targeting the G4 Hot-Spot.
Chemistry, 2020
|
|
8IQI
 
 | | Structure of Full-Length AsfvPrimPol in Complex-Form | | 分子名称: | DNA (32-MER), MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | 著者 | Shao, Z.W, Su, S.C, Gan, J.H. | | 登録日 | 2023-03-16 | | 公開日 | 2023-07-26 | | 最終更新日 | 2023-11-29 | | 実験手法 | ELECTRON MICROSCOPY (3.32 Å) | | 主引用文献 | Structures and implications of the C962R protein of African swine fever virus.
Nucleic Acids Res., 51, 2023
|
|
8I4V
 
 | | Cryo-EM structure of 5-subunit Smc5/6 arm region | | 分子名称: | DNA repair protein KRE29, E3 SUMO-protein ligase MMS21, Structural maintenance of chromosomes protein 5, ... | | 著者 | Qian, L, Jun, Z, Xiang, Z, Cheng, T, Zhaoning, W, Zhenguo, C, Wang, L. | | 登録日 | 2023-01-21 | | 公開日 | 2024-06-26 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (5.97 Å) | | 主引用文献 | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 2024
|
|
8I4W
 
 | | Cryo-EM structure of 5-subunit Smc5/6 head region | | 分子名称: | DNA repair protein KRE29, Non-structural maintenance of chromosome element 5, Structural maintenance of chromosomes protein 5, ... | | 著者 | Qian, L, Jun, Z, Xiang, Z, Zhaoning, W, Cheng, T, Duo, J, Zhenguo, C, Wang, L. | | 登録日 | 2023-01-21 | | 公開日 | 2024-06-26 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (6.01 Å) | | 主引用文献 | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 2024
|
|
419D
 
 | |
8BQY
 
 | | An i-motif domain able to undergo pH-dependent conformational transitions (acidic structure) | | 分子名称: | DNA (5'-D(*CP*(DNR)P*GP*TP*TP*(DNR)P*(DNR)P*GP*TP*TP*TP*TP*TP*CP*CP*GP*TP*TP*(DNR)P*CP*GP*T)-3') | | 著者 | Serrano-Chacon, I, Mir, B, Cupellini, L, Colizzi, F, Orozco, M, Escaja, N, Gonzalez, C. | | 登録日 | 2022-11-22 | | 公開日 | 2023-02-22 | | 最終更新日 | 2024-06-19 | | 実験手法 | SOLUTION NMR | | 主引用文献 | pH-Dependent Capping Interactions Induce Large-Scale Structural Transitions in i-Motifs.
J.Am.Chem.Soc., 145, 2023
|
|
3WVG
 
 | | Time-Resolved Crystal Structure of HindIII with 0sec soaking | | 分子名称: | DNA (5'-D(*GP*CP*CP*AP*AP*GP*CP*TP*TP*GP*GP*C)-3'), GLYCEROL, SODIUM ION, ... | | 著者 | Kawamura, T, Kobayashi, T, Watanabe, N. | | 登録日 | 2014-05-21 | | 公開日 | 2015-04-08 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Analysis of the HindIII-catalyzed reaction by time-resolved crystallography
Acta Crystallogr.,Sect.D, 71, 2015
|
|
2L5U
 
 | |
4CGY
 
 | |
8HQS
 
 | | Cryo-EM structure of 8-subunit Smc5/6 head region | | 分子名称: | DNA repair protein KRE29, Non-structural maintenance of chromosome element 3, Non-structural maintenance of chromosome element 4, ... | | 著者 | Qian, L, Jun, Z, Xiang, Z, Tong, C, Wang, Z, Duo, J, Zhenguo, C, Wang, L. | | 登録日 | 2022-12-14 | | 公開日 | 2024-06-19 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 2024
|
|
7VBM
 
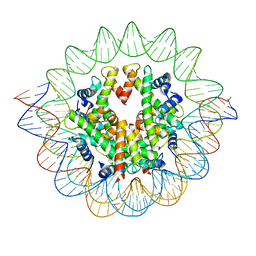 | | The mouse nucleosome structure containing H3mm18 aided by PL2-6 scFv | | 分子名称: | DNA (126-MER), Histone H2A type 1-B, Histone H2B type 3-A, ... | | 著者 | Hirai, S, Takizawa, Y, Kujirai, T, Kurumizaka, H. | | 登録日 | 2021-08-31 | | 公開日 | 2022-01-19 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Unusual nucleosome formation and transcriptome influence by the histone H3mm18 variant.
Nucleic Acids Res., 50, 2022
|
|
1SZP
 
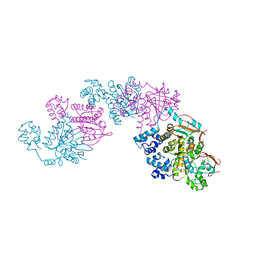 | | A Crystal Structure of the Rad51 Filament | | 分子名称: | DNA repair protein RAD51, SULFATE ION | | 著者 | Conway, A.B, Lynch, T.W, Zhang, Y, Fortin, G.S, Symington, L.S, Rice, P.A. | | 登録日 | 2004-04-06 | | 公開日 | 2004-07-13 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (3.25 Å) | | 主引用文献 | Crystal structure of a Rad51 filament.
Nat.Struct.Mol.Biol., 11, 2004
|
|
