2LPT
 
 | |
6UHW
 
 | |
4AYK
 
 | | CATALYTIC FRAGMENT OF HUMAN FIBROBLAST COLLAGENASE COMPLEXED WITH CGS-27023A, NMR, 30 STRUCTURES | | 分子名称: | CALCIUM ION, N-HYDROXY-2(R)-[[(4-METHOXYPHENYL)SULFONYL](3-PICOLYL)AMINO]-3-METHYLBUTANAMIDE HYDROCHLORIDE, PROTEIN (COLLAGENASE), ... | | 著者 | Powers, R, Moy, F.J. | | 登録日 | 1999-02-01 | | 公開日 | 1999-06-05 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR solution structure of the catalytic fragment of human fibroblast collagenase complexed with a sulfonamide derivative of a hydroxamic acid compound.
Biochemistry, 38, 1999
|
|
3AYK
 
 | | CATALYTIC FRAGMENT OF HUMAN FIBROBLAST COLLAGENASE COMPLEXED WITH CGS-27023A, NMR, MINIMIZED AVERAGE STRUCTURE | | 分子名称: | CALCIUM ION, N-HYDROXY-2(R)-[[(4-METHOXYPHENYL)SULFONYL](3-PICOLYL)AMINO]-3-METHYLBUTANAMIDE HYDROCHLORIDE, PROTEIN (COLLAGENASE), ... | | 著者 | Powers, R, Moy, F.J. | | 登録日 | 1999-02-01 | | 公開日 | 1999-06-07 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR solution structure of the catalytic fragment of human fibroblast collagenase complexed with a sulfonamide derivative of a hydroxamic acid compound.
Biochemistry, 38, 1999
|
|
1MVG
 
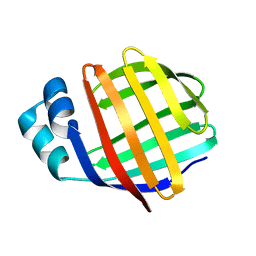 | | NMR solution structure of chicken Liver basic Fatty Acid Binding Protein (Lb-FABP) | | 分子名称: | Liver basic Fatty Acid Binding Protein | | 著者 | Vasile, F, Ragona, L, Catalano, M, Zetta, L, Perduca, M, Monaco, H, Molinari, H. | | 登録日 | 2002-09-25 | | 公開日 | 2003-03-04 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution Structure of chicken Liver basic type Fatty Acid Binding Protein
J.BIOMOL.NMR, 25, 2003
|
|
1EZY
 
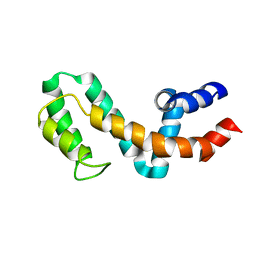 | | HIGH-RESOLUTION SOLUTION STRUCTURE OF FREE RGS4 BY NMR | | 分子名称: | REGULATOR OF G-PROTEIN SIGNALING 4 | | 著者 | Moy, F.J, Chanda, P.K, Cockett, M.I, Edris, W, Jones, P.G, Mason, K, Semus, S, Powers, R. | | 登録日 | 2000-05-12 | | 公開日 | 2001-01-15 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR structure of free RGS4 reveals an induced conformational change upon binding Galpha.
Biochemistry, 39, 2000
|
|
1EZT
 
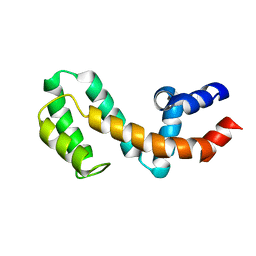 | | HIGH-RESOLUTION SOLUTION STRUCTURE OF FREE RGS4 BY NMR | | 分子名称: | REGULATOR OF G-PROTEIN SIGNALING 4 | | 著者 | Moy, F.J, Chanda, P.K, Cockett, M.I, Edris, W, Jones, P.G, Mason, K, Semus, S, Powers, R. | | 登録日 | 2000-05-11 | | 公開日 | 2001-01-15 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR structure of free RGS4 reveals an induced conformational change upon binding Galpha.
Biochemistry, 39, 2000
|
|
1BF8
 
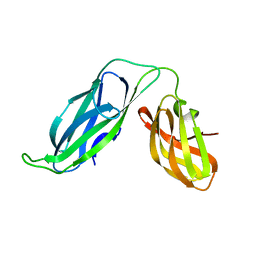 | | PERIPLASMIC CHAPERONE FIMC, NMR, 20 STRUCTURES | | 分子名称: | CHAPERONE PROTEIN FIMC | | 著者 | Pellecchia, M, Guntert, P, Glockshuber, R, Wuthrich, K. | | 登録日 | 1998-05-28 | | 公開日 | 1998-11-18 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR solution structure of the periplasmic chaperone FimC.
Nat.Struct.Biol., 5, 1998
|
|
1DE2
 
 | |
1DE1
 
 | |
1EKW
 
 | | NMR STRUCTURE OF A DNA THREE-WAY JUNCTION | | 分子名称: | DNA (5'-D(*CP*GP*GP*TP*GP*CP*GP*TP*CP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*CP*CP*AP*CP*CP*G)-3'), DNA (5'-D(*GP*GP*AP*CP*GP*TP*CP*GP*CP*AP*GP*C)-3') | | 著者 | Thiviyanathan, V, Luxon, B.A, Leontis, N.B, Donne, D, Gorenstein, D.G. | | 登録日 | 2000-03-09 | | 公開日 | 2000-03-20 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Hybrid-hybrid matrix structural refinement of a DNA three-way junction from 3D NOESY-NOESY.
J.Biomol.NMR, 14, 1999
|
|
5T82
 
 | |
1S6L
 
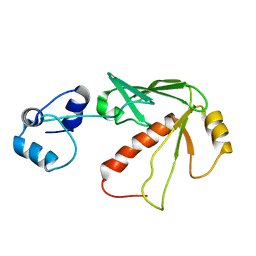 | | Solution structure of MerB, the Organomercurial Lyase involved in the bacterial mercury resistance system | | 分子名称: | Alkylmercury lyase | | 著者 | Di Lello, P, Benison, G.C, Valafar, H, Pitts, K.E, Summers, A.O, Legault, P, Omichinski, J.G. | | 登録日 | 2004-01-25 | | 公開日 | 2005-04-19 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR structural studies reveal a novel protein fold for MerB, the organomercurial lyase involved in the bacterial mercury resistance system.
Biochemistry, 43, 2004
|
|
1WCJ
 
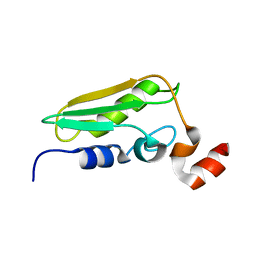 | | Conserved Hypothetical Protein TM0487 from Thermotoga maritima | | 分子名称: | HYPOTHETICAL PROTEIN TM0487 | | 著者 | Almeida, M.S, Peti, W, Herrmann, T, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | 登録日 | 2004-11-17 | | 公開日 | 2004-12-14 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR Structure of the Conserved Hypothetical Protein Tm0487 from Thermotoga Maritima: Implications for 216 Homologous Duf59 Proteins.
Protein Sci., 14, 2005
|
|
2DCP
 
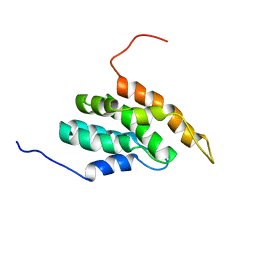 | |
2JSE
 
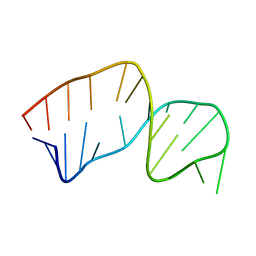 | |
2DCQ
 
 | |
2JV1
 
 | |
2LPN
 
 | | Solution Structure of N-Terminal domain of human Conserved Dopamine Neurotrophic Factor (CDNF) | | 分子名称: | Cerebral dopamine neurotrophic factor | | 著者 | Latge, C, Cabral, K.M.S, Foguel, D, Pires, J.R.M, Almeida, M.S. | | 登録日 | 2012-02-15 | | 公開日 | 2013-02-20 | | 最終更新日 | 2013-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | (1)H-, (13)C- and (15)N-NMR assignment of the N-terminal domain of human cerebral dopamine neurotrophic factor (CDNF).
Biomol.Nmr Assign., 7, 2013
|
|
2MMV
 
 | |
3IFB
 
 | | NMR STUDY OF HUMAN INTESTINAL FATTY ACID BINDING PROTEIN | | 分子名称: | INTESTINAL FATTY ACID BINDING PROTEIN | | 著者 | Zhang, F, Luecke, C, Baier, L.J, Sacchettini, J.C, Hamilton, J.A. | | 登録日 | 1998-10-16 | | 公開日 | 1998-10-21 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of human intestinal fatty acid binding protein: implications for ligand entry and exit.
J.Biomol.NMR, 9, 1997
|
|
1RON
 
 | |
2JQ2
 
 | |
8DFZ
 
 | |
8VSX
 
 | | NMR Structure of GCAP5 R22A | | 分子名称: | Guanylyl cyclase-activating protein 1 | | 著者 | Cudia, D.L, Ames, J.B. | | 登録日 | 2024-01-24 | | 公開日 | 2024-05-08 | | 最終更新日 | 2024-06-05 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR Structure of Retinal Guanylate Cyclase Activating Protein 5 (GCAP5) with R22A Mutation That Abolishes Dimerization and Enhances Cyclase Activation.
Biochemistry, 63, 2024
|
|
