3MRI
 
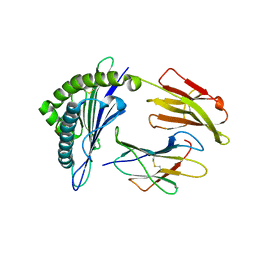 | | Crystal Structure of MHC class I HLA-A2 molecule complexed with HCV NS3-1073-1081 nonapeptide G4M-V5W variant | | 分子名称: | 9-meric peptide from Serine protease/NTPase/helicase NS3, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | 著者 | Reiser, J.-B, Le Gorrec, M, Chouquet, A, Debeaupuis, E, Echasserieau, K, Saulquin, X, Bonneville, M, Housset, D. | | 登録日 | 2010-04-29 | | 公開日 | 2011-05-25 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal Structure of MHC class I HLA-A2 molecule complexed with HCV NS3-1073-1081 nonapeptide G4M-V5W variant
To be Published
|
|
7L7A
 
 | | Solution Structure of NuxVA | | 分子名称: | NuxVA | | 著者 | Mari, F, Dovell, S. | | 登録日 | 2020-12-28 | | 公開日 | 2021-01-13 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Conus venom fractions inhibit the adhesion of Plasmodium falciparum erythrocyte membrane protein 1 domains to the host vascular receptors.
J Proteomics, 234, 2020
|
|
3DDY
 
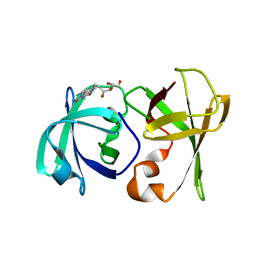 | | Structure of lumazine protein, an optical transponder of luminescent bacteria | | 分子名称: | Lumazine protein, RIBOFLAVIN | | 著者 | Chatwell, L, Illarionova, V, Illarionov, B, Skerra, A, Bacher, A, Fischer, M. | | 登録日 | 2008-06-07 | | 公開日 | 2008-07-01 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structure of lumazine protein, an optical transponder of luminescent bacteria.
J.Mol.Biol., 382, 2008
|
|
3MRN
 
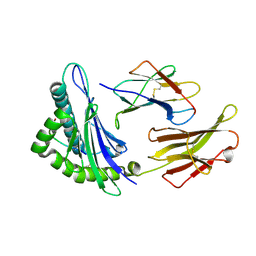 | | Crystal Structure of MHC class I HLA-A2 molecule complexed with HCV NS4b-1807-1816 decapeptide | | 分子名称: | 10-meric peptide from Genome polyprotein, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | 著者 | Gras, S, Chouquet, A, Echasserieau, K, Saulquin, X, Bonneville, M, Housset, D. | | 登録日 | 2010-04-29 | | 公開日 | 2011-05-25 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal Structure of MHC class I HLA-A2 molecule complexed with HCV NS4b-1807-1816 decapeptide
To be Published
|
|
4PQH
 
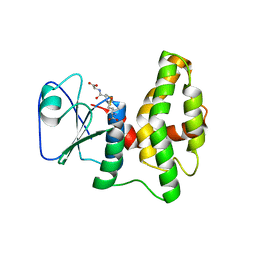 | | Crystal structure of glutathione transferase lambda1 from Populus trichocarpa | | 分子名称: | GLUTATHIONE, SODIUM ION, glutathione transferase lambda1 | | 著者 | Lallement, P.A, Meux, E, Gualberto, J.M, Prosper, P, Didierjean, C, Haouz, A, Saul, F, Rouhier, N, Hecker, A. | | 登録日 | 2014-03-03 | | 公開日 | 2014-06-25 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structural and enzymatic insights into Lambda glutathione transferases from Populus trichocarpa, monomeric enzymes constituting an early divergent class specific to terrestrial plants.
Biochem.J., 462, 2014
|
|
3DEH
 
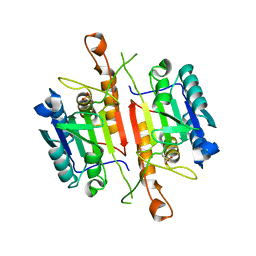 | | Crystal Structures of Caspase-3 with Bound Isoquinoline-1,3,4-trione Derivative Inhibitors | | 分子名称: | Caspase-3, isoquinoline-1,3,4(2H)-trione | | 著者 | Wu, J, Du, J, Li, J, Ding, J. | | 登録日 | 2008-06-10 | | 公開日 | 2008-09-02 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Isoquinoline-1,3,4-trione Derivatives Inactivate Caspase-3 by Generation of Reactive Oxygen Species
J.Biol.Chem., 283, 2008
|
|
8IDU
 
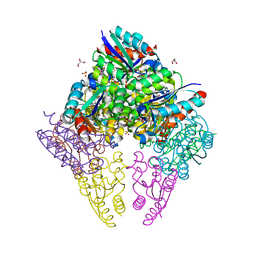 | | Crystal structure of substrate bound-form dehydroquinate dehydratase from Corynebacterium glutamicum | | 分子名称: | 1,3,4-TRIHYDROXY-5-OXO-CYCLOHEXANECARBOXYLIC ACID, 3-dehydroquinate dehydratase, GLYCEROL, ... | | 著者 | Lee, C.H, Kim, S, Kim, K.-J. | | 登録日 | 2023-02-14 | | 公開日 | 2023-12-27 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural and Biochemical Analysis of 3-Dehydroquinate Dehydratase from Corynebacterium glutamicum .
J Microbiol Biotechnol., 33, 2023
|
|
7S3B
 
 | |
5Y9Q
 
 | |
3DDH
 
 | | The structure of a putative haloacid dehalogenase-like family hydrolase from Bacteroides thetaiotaomicron VPI-5482 | | 分子名称: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | 著者 | Cuff, M.E, Duggan, E, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2008-06-05 | | 公開日 | 2008-09-30 | | 最終更新日 | 2017-10-25 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The structure of a putative haloacid dehalogenase-like family hydrolase from Bacteroides thetaiotaomicron VPI-5482
TO BE PUBLISHED
|
|
7S3C
 
 | |
3MQH
 
 | | crystal structure of the 3-N-acetyl transferase WlbB from Bordetella petrii in complex with CoA and UDP-3-amino-2-acetamido-2,3-dideoxy glucuronic acid | | 分子名称: | (2S,3S,4R,5R,6R)-5-(acetylamino)-4-amino-6-{[(R)-{[(R)-{[(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methoxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]oxy}-3-hydroxytetrahydro-2H-pyran-2-carboxylic acid, 1,2-ETHANEDIOL, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, ... | | 著者 | thoden, J.B, holden, H.M. | | 登録日 | 2010-04-28 | | 公開日 | 2010-05-12 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.43 Å) | | 主引用文献 | Molecular structure of WlbB, a bacterial N-acetyltransferase involved in the biosynthesis of 2,3-diacetamido-2,3-dideoxy-D-mannuronic acid .
Biochemistry, 49, 2010
|
|
5M43
 
 | |
3M0J
 
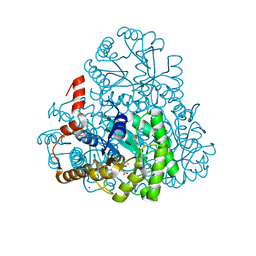 | | Structure of oxaloacetate acetylhydrolase in complex with the inhibitor 3,3-difluorooxalacetate | | 分子名称: | 2,2-difluoro-3,3-dihydroxybutanedioic acid, CALCIUM ION, MANGANESE (II) ION, ... | | 著者 | Herzberg, O, Chen, C. | | 登録日 | 2010-03-03 | | 公開日 | 2010-06-16 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Structure of oxalacetate acetylhydrolase, a virulence factor of the chestnut blight fungus.
J.Biol.Chem., 285, 2010
|
|
8HYN
 
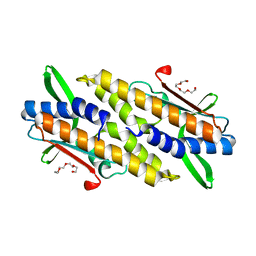 | | Bacterial STING from Riemerella anatipestifer | | 分子名称: | CD-NTase-associated protein 12, TETRAETHYLENE GLYCOL | | 著者 | Wang, Y.-C, Yang, C.-S, Hou, M.-H, Chen, Y. | | 登録日 | 2023-01-07 | | 公開日 | 2024-01-10 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.089 Å) | | 主引用文献 | Structural insights into the regulation, ligand recognition, and oligomerization of bacterial STING.
Nat Commun, 14, 2023
|
|
3MRB
 
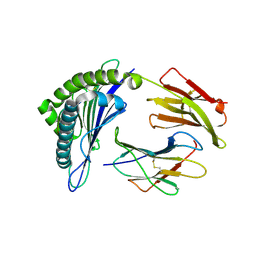 | | Crystal Structure of MHC class I HLA-A2 molecule complexed with HCMV pp65-495-503 nonapeptide A7H variant | | 分子名称: | 9-meric peptide from Tegument protein pp65, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | 著者 | Gras, S, Reiser, J.-B, Chouquet, A, Debeaupuis, E, Echasserieau, K, Saulquin, X, Bonneville, M, Housset, D. | | 登録日 | 2010-04-29 | | 公開日 | 2011-05-25 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Crystal Structure of MHC class I HLA-A2 molecule complexed with short peptide
To be Published
|
|
3DEQ
 
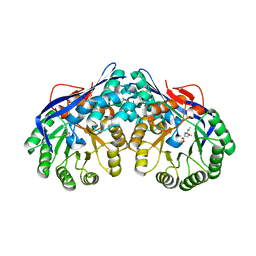 | | Crystal structure of dipeptide epimerase from Thermotoga maritima complexed with L-Ala-L-Leu dipeptide | | 分子名称: | ALANINE, LEUCINE, MAGNESIUM ION, ... | | 著者 | Fedorov, A.A, Fedorov, E.V, Imker, H.J, Gerlt, J.A, Almo, S.C. | | 登録日 | 2008-06-10 | | 公開日 | 2008-11-25 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Discovery of a dipeptide epimerase enzymatic function guided by homology modeling and virtual screening.
Structure, 16, 2008
|
|
8IDR
 
 | |
7S3A
 
 | |
4PFP
 
 | | Myosin VI motor domain in the Pi release state (with Pi) space group P21 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | 著者 | Isabet, T, Benisty, H, Llinas, P, Sweeney, H.L, Houdusse, A. | | 登録日 | 2014-04-30 | | 公開日 | 2015-04-29 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.32 Å) | | 主引用文献 | How actin initiates the motor activity of Myosin.
Dev.Cell, 33, 2015
|
|
3DGO
 
 | | A non-biological ATP binding protein with a Tyr-Phe mutation in the ligand binding domain | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, ATP Binding Protein-DX, CHLORIDE ION, ... | | 著者 | Simmons, C.R, Allen, J.P, Chaput, J.C. | | 登録日 | 2008-06-13 | | 公開日 | 2009-06-30 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | A synthetic protein selected for ligand binding affinity mediates ATP hydrolysis.
Acs Chem.Biol., 4, 2009
|
|
1F7A
 
 | |
8C3T
 
 | |
8HWJ
 
 | | Bacterial STING from Epilithonimonas lactis in complex with 3'3'-c-di-AMP | | 分子名称: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, CD-NTase-associated protein 12 | | 著者 | Wang, Y.-C, Yang, C.-S, Hou, M.-H, Chen, Y. | | 登録日 | 2022-12-30 | | 公開日 | 2024-01-03 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.553 Å) | | 主引用文献 | Structural insights into the regulation, ligand recognition, and oligomerization of bacterial STING.
Nat Commun, 14, 2023
|
|
3SXK
 
 | |
