8BTR
 
 | | Giardia Ribosome in PRE-T Hybrid State (D2) | | 分子名称: | 5.8S rRNA, 5S rRNA, Large Subunit rRNA, ... | | 著者 | Majumdar, S, Emmerich, A.G, Sanyal, S. | | 登録日 | 2022-11-29 | | 公開日 | 2023-03-22 | | 最終更新日 | 2023-05-03 | | 実験手法 | ELECTRON MICROSCOPY (3.25 Å) | | 主引用文献 | Insights into translocation mechanism and ribosome evolution from cryo-EM structures of translocation intermediates of Giardia intestinalis.
Nucleic Acids Res., 51, 2023
|
|
8BR8
 
 | | Giardia ribosome in POST-T state (A1) | | 分子名称: | 40S ribosomal protein S21, 40S ribosomal protein S25, 40S ribosomal protein S26, ... | | 著者 | Majumdar, S, Emmerich, A.G, Sanyal, S. | | 登録日 | 2022-11-22 | | 公開日 | 2023-03-15 | | 最終更新日 | 2023-05-03 | | 実験手法 | ELECTRON MICROSCOPY (3.35 Å) | | 主引用文献 | Insights into translocation mechanism and ribosome evolution from cryo-EM structures of translocation intermediates of Giardia intestinalis.
Nucleic Acids Res., 51, 2023
|
|
8BRM
 
 | | Giardia ribosome in POST-T state, no E-site tRNA (A6) | | 分子名称: | 5.8S rRNA, 5S rRNA, Large Subunit rRNA, ... | | 著者 | Majumdar, S, Emmerich, A.G, Sanyal, S. | | 登録日 | 2022-11-23 | | 公開日 | 2023-03-15 | | 最終更新日 | 2023-05-03 | | 実験手法 | ELECTRON MICROSCOPY (3.33 Å) | | 主引用文献 | Insights into translocation mechanism and ribosome evolution from cryo-EM structures of translocation intermediates of Giardia intestinalis.
Nucleic Acids Res., 51, 2023
|
|
1EFK
 
 | | STRUCTURE OF HUMAN MALIC ENZYME IN COMPLEX WITH KETOMALONATE | | 分子名称: | ALPHA-KETOMALONIC ACID, MAGNESIUM ION, MALIC ENZYME, ... | | 著者 | Yang, Z, Floyd, D.L, Loeber, G, Tong, L. | | 登録日 | 2000-02-09 | | 公開日 | 2000-03-08 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structure of a closed form of human malic enzyme and implications for catalytic mechanism.
Nat.Struct.Biol., 7, 2000
|
|
8BSI
 
 | | Giardia ribosome chimeric hybrid-like GDP+Pi bound state (B1) | | 分子名称: | 40S ribosomal protein S21, 40S ribosomal protein S25, 40S ribosomal protein S26, ... | | 著者 | Majumdar, S, Emmerich, A.G, Sanyal, S. | | 登録日 | 2022-11-25 | | 公開日 | 2023-03-15 | | 最終更新日 | 2023-05-03 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Insights into translocation mechanism and ribosome evolution from cryo-EM structures of translocation intermediates of Giardia intestinalis.
Nucleic Acids Res., 51, 2023
|
|
4B1V
 
 | | Structure of the Phactr1 RPEL-N domain bound to G-actin | | 分子名称: | 1,2-ETHANEDIOL, ACTIN, ALPHA SKELETAL MUSCLE, ... | | 著者 | Mouilleron, S, Wiezlak, M, O'Reilly, N, Treisman, R, McDonald, N.Q. | | 登録日 | 2012-07-12 | | 公開日 | 2012-11-07 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structures of the Phactr1 RPEL domain and RPEL motif complexes with G-actin reveal the molecular basis for actin binding cooperativity.
Structure, 20, 2012
|
|
4B1W
 
 | | Structure of the Phactr1 RPEL-2 domain bound to actin | | 分子名称: | ACTIN, ALPHA SKELETAL MUSCLE, ADENOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Mouilleron, S, Wiezlak, M, O'Reilly, N, Treisman, R, McDonald, N.Q. | | 登録日 | 2012-07-12 | | 公開日 | 2013-07-31 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structures of the Phactr1 RPEL domain and RPEL motif complexes with G-actin reveal the molecular basis for actin binding cooperativity.
Structure, 20, 2012
|
|
1EPH
 
 | |
6XIR
 
 | | Cryo-EM Structure of K63 Ubiquitinated Yeast Translocating Ribosome under Oxidative Stress | | 分子名称: | 18S ribosomal RNA, 35S ribosomal RNA, 40S ribosomal protein S0-A, ... | | 著者 | Zhou, Y, Bartesaghi, A, Silva, G.M. | | 登録日 | 2020-06-21 | | 公開日 | 2020-08-26 | | 最終更新日 | 2020-09-23 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structural impact of K63 ubiquitin on yeast translocating ribosomes under oxidative stress.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4B1X
 
 | | Structure of the Phactr1 RPEL-2 bound to G-actin | | 分子名称: | ACTIN, ALPHA SKELETAL MUSCLE, ADENOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Mouilleron, S, Wiezlak, M, O'Reilly, N, Treisman, R, McDonald, N.Q. | | 登録日 | 2012-07-12 | | 公開日 | 2013-07-31 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structures of the Phactr1 RPEL domain and RPEL motif complexes with G-actin reveal the molecular basis for actin binding cooperativity.
Structure, 20, 2012
|
|
1RW9
 
 | | Crystal structure of the Arthrobacter aurescens chondroitin AC lyase | | 分子名称: | PHOSPHATE ION, SODIUM ION, chondroitin AC lyase | | 著者 | Lunin, V.V, Li, Y, Linhardt, R.J, Miyazono, H, Kyogashima, M, Kaneko, T, Bell, A.W, Cygler, M. | | 登録日 | 2003-12-16 | | 公開日 | 2004-04-13 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | High-resolution crystal structure of Arthrobacter aurescens chondroitin AC lyase: an enzyme-substrate complex defines the catalytic mechanism
J.Mol.Biol., 337, 2004
|
|
1E5C
 
 | | Internal xylan binding domain from C. fimi Xyn10A, R262G mutant | | 分子名称: | XYLANASE D | | 著者 | Simpson, P.J, Hefang, X, Bolam, D.N, Gilbert, H.J, Williamson, M.P. | | 登録日 | 2000-07-24 | | 公開日 | 2001-05-25 | | 最終更新日 | 2018-10-24 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The Structural Basis for the Ligand Specificity of Family 2 Carbohydrate Binding Nodules
J.Biol.Chem., 275, 2000
|
|
1ESY
 
 | |
5ADH
 
 | |
4B1Y
 
 | | Structure of the Phactr1 RPEL-3 bound to G-actin | | 分子名称: | ACTIN, ALPHA SKELETAL MUSCLE, ADENOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Mouilleron, S, Wiezlak, M, O'Reilly, N, Treisman, R, McDonald, N.Q. | | 登録日 | 2012-07-12 | | 公開日 | 2013-07-31 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.29 Å) | | 主引用文献 | Structures of the Phactr1 RPEL domain and RPEL motif complexes with G-actin reveal the molecular basis for actin binding cooperativity.
Structure, 20, 2012
|
|
1RWF
 
 | | Crystal structure of Arthrobacter aurescens chondroitin AC lyase in complex with chondroitin tetrasaccharide | | 分子名称: | 2,6-anhydro-3-deoxy-L-threo-hex-2-enonic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-2,6-anhydro-3-deoxy-L-xylo-hexonic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose, PHOSPHATE ION, SODIUM ION, ... | | 著者 | Lunin, V.V, Li, Y, Miyazono, H, Kyogashima, M, Bell, A.W, Cygler, M. | | 登録日 | 2003-12-16 | | 公開日 | 2004-04-13 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | High-resolution crystal structure of Arthrobacter aurescens chondroitin AC lyase: an enzyme-substrate complex defines the catalytic mechanism
J.Mol.Biol., 337, 2004
|
|
1RWG
 
 | | Crystal structure of Arthrobacter aurescens chondroitin AC lyase in complex with chondroitin tetrasaccharide | | 分子名称: | 2,6-anhydro-3-deoxy-L-threo-hex-2-enonic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-2,6-anhydro-3-deoxy-L-xylo-hexonic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose, PHOSPHATE ION, SODIUM ION, ... | | 著者 | Lunin, V.V, Li, Y, Miyazono, H, Kyogashima, M, Bell, A.W, Cygler, M. | | 登録日 | 2003-12-16 | | 公開日 | 2004-04-13 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | High-resolution crystal structure of Arthrobacter aurescens chondroitin AC lyase: an enzyme-substrate complex defines the catalytic mechanism
J.Mol.Biol., 337, 2004
|
|
8EVT
 
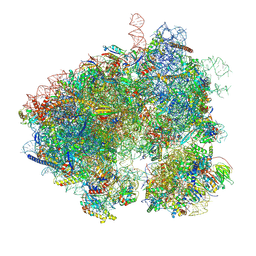 | | Hypopseudouridylated yeast 80S bound with Taura syndrome virus (TSV) internal ribosome entry site (IRES) refined against a composite map | | 分子名称: | 18S rRNA, 25S rRNA, 40S ribosomal protein S0-A, ... | | 著者 | Zhao, Y, Rai, J, Li, H. | | 登録日 | 2022-10-20 | | 公開日 | 2023-09-06 | | 最終更新日 | 2023-11-01 | | 実験手法 | ELECTRON MICROSCOPY (2.2 Å) | | 主引用文献 | Regulation of translation by ribosomal RNA pseudouridylation.
Sci Adv, 9, 2023
|
|
5MQF
 
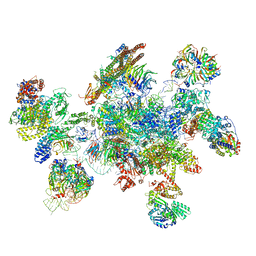 | | Cryo-EM structure of a human spliceosome activated for step 2 of splicing (C* complex) | | 分子名称: | 116 kDa U5 small nuclear ribonucleoprotein component, ATP-dependent RNA helicase DHX8, Cell division cycle 5-like protein, ... | | 著者 | Bertram, K, Hartmuth, K, Kastner, B. | | 登録日 | 2016-12-20 | | 公開日 | 2017-03-22 | | 最終更新日 | 2018-11-21 | | 実験手法 | ELECTRON MICROSCOPY (5.9 Å) | | 主引用文献 | Cryo-EM structure of a human spliceosome activated for step 2 of splicing.
Nature, 542, 2017
|
|
5XY3
 
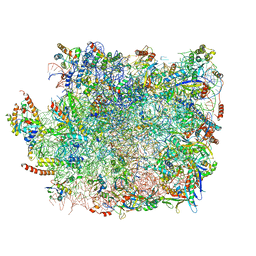 | | Large subunit of Trichomonas vaginalis ribosome | | 分子名称: | 25S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | 著者 | Li, Z, Guo, Q, Zheng, L, Ji, Y, Xie, Y, Lai, D, Lun, Z, Suo, X, Gao, N. | | 登録日 | 2017-07-06 | | 公開日 | 2017-08-30 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Cryo-EM structures of the 80S ribosomes from human parasites Trichomonas vaginalis and Toxoplasma gondii
Cell Res., 27, 2017
|
|
6XIQ
 
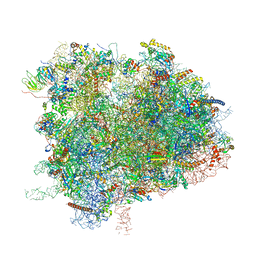 | | Cryo-EM Structure of K63R Ubiquitin Mutant Ribosome under Oxidative Stress | | 分子名称: | 18S ribosomal RNA, 35S ribosomal RNA, 40S ribosomal protein S0-A, ... | | 著者 | Zhou, Y, Bartesaghi, A, Silva, G.M. | | 登録日 | 2020-06-21 | | 公開日 | 2020-08-26 | | 最終更新日 | 2020-09-23 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Structural impact of K63 ubiquitin on yeast translocating ribosomes under oxidative stress.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8EVP
 
 | | Hypopseudouridylated yeast 80S bound with Taura syndrome virus (TSV) internal ribosome entry site (IRES), Structure I | | 分子名称: | 18S rRNA, 25S rRNA, 40S ribosomal protein S0-A, ... | | 著者 | Zhao, Y, Rai, J, Li, H. | | 登録日 | 2022-10-20 | | 公開日 | 2023-09-06 | | 実験手法 | ELECTRON MICROSCOPY (2.38 Å) | | 主引用文献 | Regulation of translation by ribosomal RNA pseudouridylation.
Sci Adv, 9, 2023
|
|
8EWC
 
 | | Hypopseudouridylated yeast 80S bound with Taura syndrome virus (TSV) internal ribosome entry site (IRES), Structure II | | 分子名称: | 18S rRNA, 25S rRNA, 40S ribosomal protein S0-A, ... | | 著者 | Zhao, Y, Rai, J, Li, H. | | 登録日 | 2022-10-22 | | 公開日 | 2023-09-06 | | 実験手法 | ELECTRON MICROSCOPY (2.45 Å) | | 主引用文献 | Regulation of translation by ribosomal RNA pseudouridylation.
Sci Adv, 9, 2023
|
|
8G83
 
 | | Structure of NAD+ consuming protein Acinetobacter baumannii TIR domain | | 分子名称: | NAD(+) hydrolase AbTIR | | 著者 | Klontz, E.H, Wang, Y, Glendening, G, Carr, J, Tsibouris, T, Buddula, S, Nallar, S, Soares, A, Snyder, G.A. | | 登録日 | 2023-02-17 | | 公開日 | 2023-10-11 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (3.03 Å) | | 主引用文献 | The structure of NAD + consuming protein Acinetobacter baumannii TIR domain shows unique kinetics and conformations.
J.Biol.Chem., 299, 2023
|
|
5YZG
 
 | | The Cryo-EM Structure of Human Catalytic Step I Spliceosome (C complex) at 4.1 angstrom resolution | | 分子名称: | 116 kDa U5 small nuclear ribonucleoprotein component, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Zhan, X, Yan, C, Zhang, X, Lei, J, Shi, Y. | | 登録日 | 2017-12-14 | | 公開日 | 2018-08-08 | | 最終更新日 | 2020-10-14 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Structure of a human catalytic step I spliceosome
Science, 359, 2018
|
|
