1M9D
 
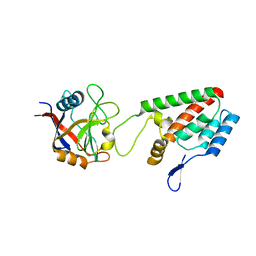 | | X-ray crystal structure of Cyclophilin A/HIV-1 CA N-terminal domain (1-146) O-type chimera Complex. | | 分子名称: | Cyclophilin A, HIV-1 Capsid | | 著者 | Howard, B.R, Vajdos, F.F, Li, S, Sundquist, W.I, Hill, C.P. | | 登録日 | 2002-07-28 | | 公開日 | 2003-05-27 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural insights into the catalytic mechanism of cyclophilin A
Nat.Struct.Biol., 10, 2003
|
|
1M9C
 
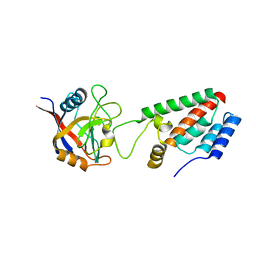 | | X-ray crystal structure of Cyclophilin A/HIV-1 CA N-terminal domain (1-146) M-type Complex. | | 分子名称: | Cyclophilin A, HIV-1 Capsid | | 著者 | Howard, B.R, Vajdos, F.F, Li, S, Sundquist, W.I, Hill, C.P. | | 登録日 | 2002-07-28 | | 公開日 | 2003-05-27 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural insights into the catalytic mechanism of cyclophilin A
Nat.Struct.Biol., 10, 2003
|
|
1M9X
 
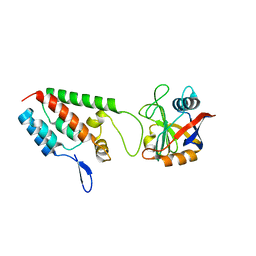 | | X-ray crystal structure of Cyclophilin A/HIV-1 CA N-terminal domain (1-146) M-type H87A,A88M,G89A Complex. | | 分子名称: | Cyclophilin A, HIV-1 Capsid | | 著者 | Howard, B.R, Vajdos, F.F, Li, S, Sundquist, W.I, Hill, C.P. | | 登録日 | 2002-07-30 | | 公開日 | 2003-05-27 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural insights into the catalytic mechanism of cyclophilin A
Nat.Struct.Biol., 10, 2003
|
|
1M9Y
 
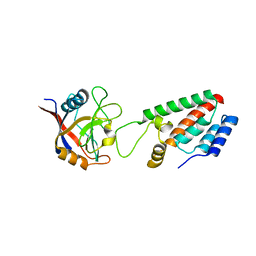 | | X-ray crystal structure of Cyclophilin A/HIV-1 CA N-terminal domain (1-146) M-type H87A,G89A Complex. | | 分子名称: | Cyclophilin A, HIV-1 Capsid | | 著者 | Howard, B.R, Vajdos, F.F, Li, S, Sundquist, W.I, Hill, C.P. | | 登録日 | 2002-07-30 | | 公開日 | 2003-05-27 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural insights into the catalytic mechanism of cyclophilin A
Nat.Struct.Biol., 10, 2003
|
|
1M9E
 
 | | X-ray crystal structure of Cyclophilin A/HIV-1 CA N-terminal domain (1-146) M-type H87A Complex. | | 分子名称: | Cyclophilin A, HIV-1 Capsid | | 著者 | Howard, B.R, Vajdos, F.F, Li, S, Sundquist, W.I, Hill, C.P. | | 登録日 | 2002-07-28 | | 公開日 | 2003-05-27 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.72 Å) | | 主引用文献 | Structural insights into the catalytic mechanism of cyclophilin A
Nat.Struct.Biol., 10, 2003
|
|
4CPM
 
 | | Structure of the Neuraminidase from the B/Brisbane/60/2008 virus in complex with Oseltamivir | | 分子名称: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Vachieri, S.G, Collins, P.J, Escuret, V, Casalegno, J.S, Cattle, N, Ferraris, O, Sabatier, M, Frobert, E, Caro, V, Skehel, J.J, Gamblin, S.J, Valla, F, Valette, M, Ottmann, M, McCauley, J.W, Daniels, R.S, Lina, B. | | 登録日 | 2014-02-07 | | 公開日 | 2014-05-14 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | A Novel I221 L Substitution in Neuraminidase Confers High Level Resistance to Oseltamivir in Influenza B Viruses.
J.Infect.Dis., 210, 2014
|
|
4CPN
 
 | | Structure of the Neuraminidase from the B/Brisbane/60/2008 virus in complex with Zanamivir | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Vachieri, S.G, Collins, P.J, Escuret, V, Casalegno, J.S, Cattle, N, Ferraris, O, Sabatier, M, Frobert, E, Caro, V, Skehel, J.J, Gamblin, S.J, Valla, F, Valette, M, Ottmann, M, McCauley, J.W, Daniels, R.S, Lina, B. | | 登録日 | 2014-02-08 | | 公開日 | 2014-05-14 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | A Novel I221 L Substitution in Neuraminidase Confers High Level Resistance to Oseltamivir in Influenza B Viruses.
J.Infect.Dis., 210, 2014
|
|
5TPB
 
 | |
7FX6
 
 | | Crystal Structure of human FABP4 in complex with N,N-diethyl-4-pyridin-4-yl-3-(1H-tetrazol-5-yl)-6,7,8,9-tetrahydro-5H-cyclohepta[b]pyridin-2-amine | | 分子名称: | (3M)-N,N-diethyl-4-(pyridin-4-yl)-3-(1H-tetrazol-5-yl)-6,7,8,9-tetrahydro-5H-cyclohepta[b]pyridin-2-amine, Fatty acid-binding protein, adipocyte, ... | | 著者 | Ehler, A, Benz, J, Obst, U, Obst-Sander, U, Rudolph, M.G. | | 登録日 | 2023-04-27 | | 公開日 | 2023-06-14 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.05 Å) | | 主引用文献 | Crystal Structure of a human FABP4 complex
To be published
|
|
7FW9
 
 | | Crystal Structure of human FABP4 in complex with 2-[(3-ethoxycarbonyl-4,5,6,7-tetrahydro-1-benzothiophen-2-yl)carbamoyl]cyclopentene-1-carboxylic acid | | 分子名称: | 2-{[3-(ethoxycarbonyl)-4,5,6,7-tetrahydro-1-benzothiophen-2-yl]carbamoyl}cyclopent-1-ene-1-carboxylic acid, Fatty acid-binding protein, adipocyte, ... | | 著者 | Ehler, A, Benz, J, Obst, U, Ceccarelli-Simona, M, Rudolph, M.G. | | 登録日 | 2023-04-27 | | 公開日 | 2023-06-14 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Crystal Structure of a human FABP4 complex
To be published
|
|
7YU1
 
 | |
7YU0
 
 | |
7YU2
 
 | |
6TC3
 
 | |
6TBV
 
 | |
5W6H
 
 | | Crystal structure of Bacteriophage CBA120 tailspike protein 4 enzymatically active domain (TSP4dN, orf213) | | 分子名称: | ACETATE ION, CHLORIDE ION, POTASSIUM ION, ... | | 著者 | Plattner, M, Shneider, M.M, Leiman, P.G. | | 登録日 | 2017-06-16 | | 公開日 | 2018-10-24 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.289 Å) | | 主引用文献 | Structure and Function of the Branched Receptor-Binding Complex of Bacteriophage CBA120.
J.Mol.Biol., 431, 2019
|
|
5W6P
 
 | | Crystal structure of Bacteriophage CBA120 tailspike protein 2 enzymatically active domain (TSP2dN, orf211) | | 分子名称: | 1,2-ETHANEDIOL, POTASSIUM ION, ZINC ION, ... | | 著者 | Plattner, M, Shneider, M.M, Leiman, P.G. | | 登録日 | 2017-06-16 | | 公開日 | 2018-10-24 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.335 Å) | | 主引用文献 | Structure and Function of the Branched Receptor-Binding Complex of Bacteriophage CBA120.
J.Mol.Biol., 431, 2019
|
|
5W6S
 
 | | Crystal structure of Bacteriophage CBA120 tailspike protein 2 enzymatically active domain (TSP2dN, orf211) complex with Escherichia Coli O157-antigen | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, POTASSIUM ION, ... | | 著者 | Plattner, M, Shneider, M.M, Leiman, P.G. | | 登録日 | 2017-06-16 | | 公開日 | 2018-10-24 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.263 Å) | | 主引用文献 | Structure and Function of the Branched Receptor-Binding Complex of Bacteriophage CBA120.
J.Mol.Biol., 431, 2019
|
|
1EIB
 
 | | CRYSTAL STRUCTURE OF CHITINASE A MUTANT D313A COMPLEXED WITH OCTA-N-ACETYLCHITOOCTAOSE (NAG)8. | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHITINASE A | | 著者 | Papanikolau, Y, Prag, G, Tavlas, G, Vorgias, C.E, Oppenheim, A.B, Petratos, K. | | 登録日 | 2000-02-25 | | 公開日 | 2001-02-25 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | High resolution structural analyses of mutant chitinase A complexes with substrates provide new insight into the mechanism of catalysis.
Biochemistry, 40, 2001
|
|
1EHN
 
 | | CRYSTAL STRUCTURE OF CHITINASE A MUTANT E315Q COMPLEXED WITH OCTA-N-ACETYLCHITOOCTAOSE (NAG)8. | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHITINASE A | | 著者 | Papanikolau, Y, Prag, G, Tavlas, G, Vorgias, C.E, Oppenheim, A.B, Petratos, K. | | 登録日 | 2000-02-22 | | 公開日 | 2001-02-22 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | High resolution structural analyses of mutant chitinase A complexes with substrates provide new insight into the mechanism of catalysis.
Biochemistry, 40, 2001
|
|
3D73
 
 | | Crystal structure of a pheromone binding protein mutant D35A, from Apis mellifera, at pH 7.0 | | 分子名称: | N-BUTYL-BENZENESULFONAMIDE, Pheromone-binding protein ASP1 | | 著者 | Pesenti, M.E, Spinelli, S, Bezirard, V, Briand, L, Pernollet, J.C, Tegoni, M, Cambillau, C. | | 登録日 | 2008-05-20 | | 公開日 | 2009-05-26 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | Queen bee pheromone binding protein pH-induced domain swapping favors pheromone release
J.Mol.Biol., 390, 2009
|
|
3D78
 
 | | Dimeric crystal structure of a pheromone binding protein mutant D35N, from apis mellifera, at pH 7.0 | | 分子名称: | 1,2-ETHANEDIOL, N-BUTYL-BENZENESULFONAMIDE, Pheromone-binding protein ASP1 | | 著者 | Pesenti, M.E, Spinelli, S, Bezirard, V, Briand, L, Pernollet, J.C, Tegoni, M, Cambillau, C. | | 登録日 | 2008-05-20 | | 公開日 | 2009-05-26 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Queen bee pheromone binding protein pH-induced domain swapping favors pheromone release
J.Mol.Biol., 390, 2009
|
|
7VNU
 
 | |
3AHS
 
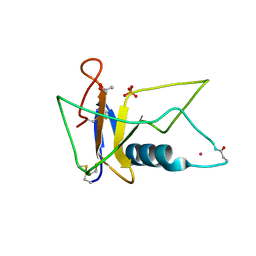 | | Crystal Structure of Ustilago sphaerogena Ribonuclease U2B | | 分子名称: | GLYCEROL, PHOSPHATE ION, POTASSIUM ION, ... | | 著者 | Noguchi, S. | | 登録日 | 2010-04-29 | | 公開日 | 2010-07-07 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.32 Å) | | 主引用文献 | Structural changes induced by the deamidation and isomerization of asparagine revealed by the crystal structure of Ustilago sphaerogena ribonuclease U2B
Biopolymers, 93, 2010
|
|
2GAE
 
 | |
