1SQ8
 
 | |
4IC1
 
 | | Crystal structure of SSO0001 | | 分子名称: | IRON/SULFUR CLUSTER, MANGANESE (II) ION, Uncharacterized protein | | 著者 | Nocek, B, Skarina, T, Lemak, S, Beloglazova, N, Flick, R, Brown, G, Savchenko, A, Joachimiak, A, Yakunin, A.F, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2012-12-09 | | 公開日 | 2013-01-16 | | 最終更新日 | 2014-07-02 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Toroidal structure and DNA cleavage by the CRISPR-associated [4Fe-4S] cluster containing Cas4 nuclease SSO0001 from Sulfolobus solfataricus.
J.Am.Chem.Soc., 135, 2013
|
|
2QTQ
 
 | |
6XJF
 
 | |
1DFA
 
 | | CRYSTAL STRUCTURE OF PI-SCEI IN C2 SPACE GROUP | | 分子名称: | PI-SCEI ENDONUCLEASE | | 著者 | Hu, D, Crist, M, Duan, X, Quiocho, F.A, Gimble, F.S. | | 登録日 | 1999-11-18 | | 公開日 | 1999-12-08 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Probing the structure of the PI-SceI-DNA complex by affinity cleavage and affinity photocross-linking.
J.Biol.Chem., 275, 2000
|
|
8HEY
 
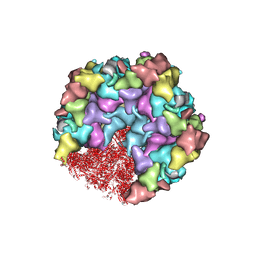 | | One CVSC-binding penton vertex in HCMV B-capsid | | 分子名称: | Capsid vertex component 1, Capsid vertex component 2, Major capsid protein, ... | | 著者 | Li, Z, Yu, X. | | 登録日 | 2022-11-09 | | 公開日 | 2023-11-01 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Cryo-electron microscopy structures of capsids and in situ portals of DNA-devoid capsids of human cytomegalovirus.
Nat Commun, 14, 2023
|
|
8HEX
 
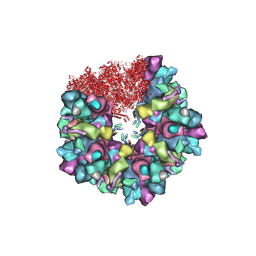 | | C5 portal vertex in HCMV B-capsid | | 分子名称: | Capsid vertex component 1, Capsid vertex component 2, Major capsid protein, ... | | 著者 | Li, Z, Yu, X. | | 登録日 | 2022-11-08 | | 公開日 | 2023-11-01 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Cryo-electron microscopy structures of capsids and in situ portals of DNA-devoid capsids of human cytomegalovirus.
Nat Commun, 14, 2023
|
|
4KIS
 
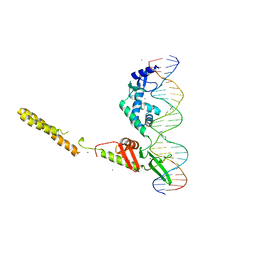 | | Crystal Structure of a LSR-DNA Complex | | 分子名称: | CALCIUM ION, DNA (26-MER), Putative integrase [Bacteriophage A118], ... | | 著者 | Rutherford, K, Yuan, P, Perry, K, Van Duyne, G.D. | | 登録日 | 2013-05-02 | | 公開日 | 2013-07-10 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Attachment site recognition and regulation of directionality by the serine integrases.
Nucleic Acids Res., 41, 2013
|
|
2MB7
 
 | |
1SD4
 
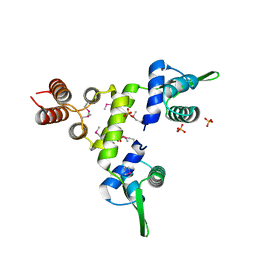 | | Crystal Structure of a SeMet derivative of BlaI at 2.0 A | | 分子名称: | PENICILLINASE REPRESSOR, SULFATE ION | | 著者 | Safo, M.K, Zhao, Q, Musayev, F.N, Robinson, H, Scarsdale, N, Archer, G.L. | | 登録日 | 2004-02-13 | | 公開日 | 2004-08-10 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structures of the BlaI repressor from Staphylococcus aureus and its complex with DNA: insights into transcriptional regulation of the bla and mec operons
J.Bacteriol., 187, 2005
|
|
1SD6
 
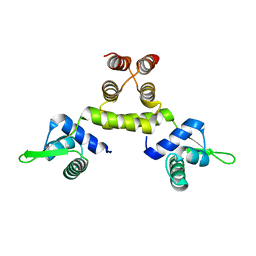 | | Crystal Structure of Native MecI at 2.65 A | | 分子名称: | Methicillin resistance regulatory protein mecI | | 著者 | Safo, M.K, Zhao, Q, Musayev, F.N, Robinson, H, Scarsdale, N, Archer, G.L. | | 登録日 | 2004-02-13 | | 公開日 | 2004-02-24 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Crystal structures of the BlaI repressor from Staphylococcus aureus and its complex with DNA: insights into transcriptional regulation of the bla and mec operons
J.Bacteriol., 187, 2005
|
|
5MMP
 
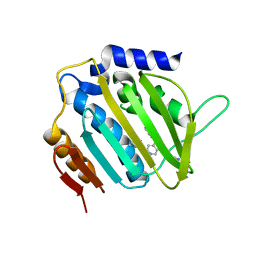 | | E. coli DNA Gyrase B 24 kDa ATPase domain in complex with 1-ethyl-3-[5-pyridin-4-yl-8-(pyridin-3-ylamino)-isoquinolin-3-yl]-urea | | 分子名称: | 1-ethyl-3-[5-pyridin-4-yl-8-(pyridin-3-ylamino)isoquinolin-3-yl]urea, DNA gyrase subunit B | | 著者 | Panchaud, P, Bruyere, T, Blumstein, A.-C, Bur, D, Chambovey, A, Ertel, E.A, Gude, M, Hubschwerlen, C, Jacob, L, Kimmerlin, T, Pfeifer, T, Prade, L, Seiler, P, Ritz, D, Rueedi, G. | | 登録日 | 2016-12-12 | | 公開日 | 2017-04-26 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Discovery and Optimization of Isoquinoline Ethyl Ureas as Antibacterial Agents.
J. Med. Chem., 60, 2017
|
|
5MMN
 
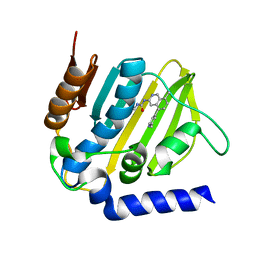 | | E. coli DNA Gyrase B 24 kDa ATPase domain in complex with 1-ethyl-3-[8-methyl-5-(2-methyl-pyridin-4-yl)-isoquinolin-3-yl]-urea | | 分子名称: | 1-ethyl-3-[8-methyl-5-(2-methylpyridin-4-yl)isoquinolin-3-yl]urea, DNA gyrase subunit B | | 著者 | Panchaud, P, Bruyere, T, Blumstein, A.-C, Bur, D, Chambovey, A, Ertel, E.A, Gude, M, Hubschwerlen, C, Jacob, L, Kimmerlin, T, Pfeifer, T, Prade, L, Seiler, P, Ritz, D, Rueedi, G. | | 登録日 | 2016-12-12 | | 公開日 | 2017-04-26 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Discovery and Optimization of Isoquinoline Ethyl Ureas as Antibacterial Agents.
J. Med. Chem., 60, 2017
|
|
7F45
 
 | | Structure of an Anti-CRISPR protein | | 分子名称: | AcrIF5 | | 著者 | Feng, Y. | | 登録日 | 2021-06-17 | | 公開日 | 2022-03-09 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.52 Å) | | 主引用文献 | AcrIF5 specifically targets DNA-bound CRISPR-Cas surveillance complex for inhibition.
Nat.Chem.Biol., 18, 2022
|
|
7NRP
 
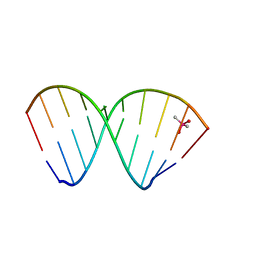 | | The crystal structure of a DNA:RNA hybrid duplex sequence CTTTTCTTTG | | 分子名称: | CACODYLATE ION, DNA (5'-D(*CP*TP*TP*TP*TP*CP*TP*TP*TP*G)-3'), RNA (5'-R(*CP*AP*AP*AP*GP*AP*AP*AP*AP*G)-3') | | 著者 | Thorpe, C, Hardwick, J, McDonough, M.A, Hall, J.P, Baker, Y.R, El-Sagheer, A.H, Brown, T. | | 登録日 | 2021-03-04 | | 公開日 | 2022-06-22 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.67 Å) | | 主引用文献 | An LNA-amide modification that enhances the cell uptake and activity of phosphorothioate exon-skipping oligonucleotides.
Nat Commun, 13, 2022
|
|
1MUS
 
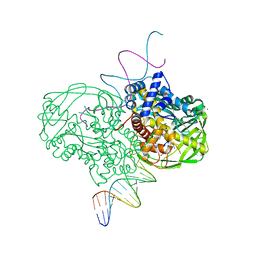 | | crystal structure of Tn5 transposase complexed with resolved outside end DNA | | 分子名称: | 1,2-ETHANEDIOL, DNA non-transferred strand, DNA transferred strand, ... | | 著者 | Holden, H.M, Thoden, J.B, Steiniger-White, M, Reznikoff, W.S, Lovell, S, Rayment, I. | | 登録日 | 2002-09-24 | | 公開日 | 2002-09-27 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure/function insights into Tn5 transposition.
Curr.Opin.Struct.Biol., 14, 2004
|
|
4FW2
 
 | |
5ZJ4
 
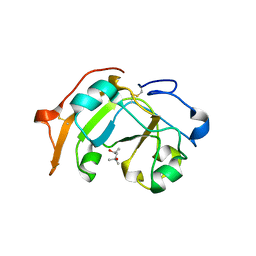 | | Guanine-specific ADP-ribosyltransferase | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, ADP-ribosyltransferase | | 著者 | Yoshida, T, Tsuge, H. | | 登録日 | 2018-03-19 | | 公開日 | 2018-08-08 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.49739277 Å) | | 主引用文献 | Substrate N2atom recognition mechanism in pierisin family DNA-targeting, guanine-specific ADP-ribosyltransferase ScARP.
J. Biol. Chem., 293, 2018
|
|
1L3A
 
 | |
8Q40
 
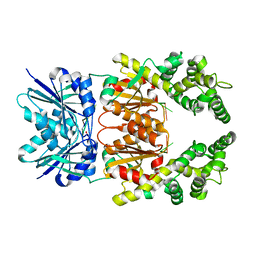 | | Crystal structure of cA4 activated Can2 in complex with a cleaved DNA substrate | | 分子名称: | Cyclic tetraadenosine monophosphate (cA4), DNA (5'-D(*TP*CP*A)-3'), DUF1887 family protein, ... | | 著者 | Jungfer, K, Sigg, A, Jinek, M. | | 登録日 | 2023-08-04 | | 公開日 | 2023-11-22 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.21 Å) | | 主引用文献 | Substrate selectivity and catalytic activation of the type III CRISPR ancillary nuclease Can2.
Nucleic Acids Res., 52, 2024
|
|
8Q42
 
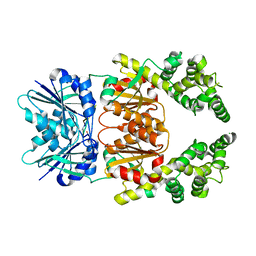 | | Crystal structure of cA4-bound Can2 (E341A) in complex with oligo-A DNA | | 分子名称: | Cyclic tetraadenosine monophosphate (cA4), DNA (5'-D(*AP*AP*AP*A)-3'), DUF1887 family protein, ... | | 著者 | Jungfer, K, Sigg, A, Jinek, M. | | 登録日 | 2023-08-04 | | 公開日 | 2023-11-22 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Substrate selectivity and catalytic activation of the type III CRISPR ancillary nuclease Can2.
Nucleic Acids Res., 52, 2024
|
|
8Q43
 
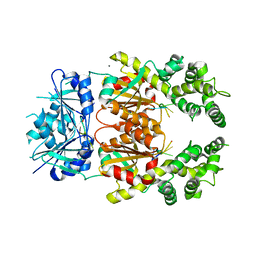 | | Crystal structure of cA4-bound Can2 (E341A) in complex with oligo-C DNA | | 分子名称: | Cyclic tetraadenosine monophosphate (cA4), DNA (5'-D(*CP*CP*CP*CP*C)-3'), DUF1887 family protein, ... | | 著者 | Jungfer, K, Sigg, A, Jinek, M. | | 登録日 | 2023-08-04 | | 公開日 | 2023-11-22 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.28 Å) | | 主引用文献 | Substrate selectivity and catalytic activation of the type III CRISPR ancillary nuclease Can2.
Nucleic Acids Res., 52, 2024
|
|
3NKD
 
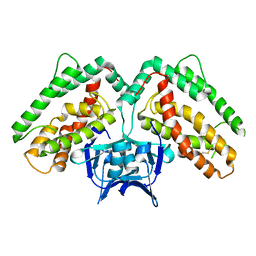 | | Structure of CRISP-associated protein Cas1 from Escherichia coli str. K-12 | | 分子名称: | CRISPR-associated protein Cas1 | | 著者 | Nocek, B, Skarina, T, Beloglazova, N, Savchenko, A, Joachimiak, A, Yakunin, A. | | 登録日 | 2010-06-18 | | 公開日 | 2010-08-25 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | A dual function of the CRISPR-Cas system in bacterial antivirus immunity and DNA repair.
Mol.Microbiol., 79, 2011
|
|
2OZE
 
 | | The Crystal structure of Delta protein of pSM19035 from Streptoccocus pyogenes | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, MAGNESIUM ION, Orf delta', ... | | 著者 | Cicek, A, Weihofen, W, Saenger, W. | | 登録日 | 2007-02-26 | | 公開日 | 2008-03-11 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.83 Å) | | 主引用文献 | Streptococcus pyogenes pSM19035 requires dynamic assembly of ATP-bound ParA and ParB on parS DNA during plasmid segregation.
Nucleic Acids Res., 36, 2008
|
|
7T31
 
 | |
