1ZIT
 
 | |
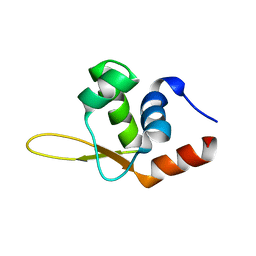
5YKL
 
 | |
5YKQ
 
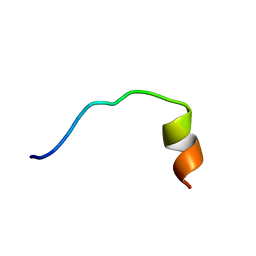 | |
5YKK
 
 | |
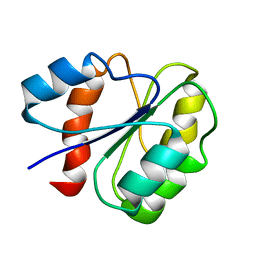
6U1O
 
 | | Structure of two-domain translational regulator Yih1 reveals a possible mechanism of action | | 分子名称: | Protein IMPACT homolog | | 著者 | Harjes, E, Jameson, G.B, Edwards, P.J.B, Goroncy, A.K, Loo, T, Norris, G.E. | | 登録日 | 2019-08-16 | | 公開日 | 2021-02-10 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Experimentally based structural model of Yih1 provides insight into its function in controlling the key translational regulator Gcn2.
Febs Lett., 595, 2021
|
|
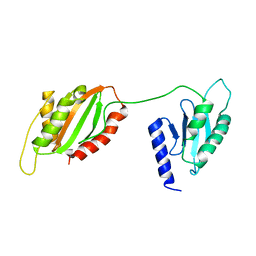
6TEY
 
 | |
6TI6
 
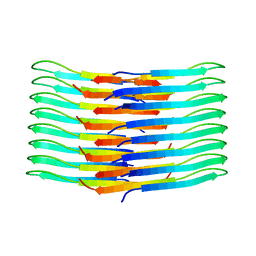 | | Mixing Abeta(1-40) and Abeta(1-42) peptides generates unique amyloid fibrils | | 分子名称: | Amyloid-beta precursor protein | | 著者 | Cerofolini, L, Ravera, E, Bologna, S, Wiglenda, T, Boddrich, A, Purfurst, B, Benilova, A, Korsak, M, Gallo, G, Rizzo, D, Gonnelli, L, Fragai, M, De Strooper, B, Wanker, E.E, Luchinat, C. | | 登録日 | 2019-11-21 | | 公開日 | 2020-07-22 | | 最終更新日 | 2024-06-19 | | 実験手法 | SOLID-STATE NMR | | 主引用文献 | Mixing A beta (1-40) and A beta (1-42) peptides generates unique amyloid fibrils.
Chem.Commun.(Camb.), 56, 2020
|
|
6TYW
 
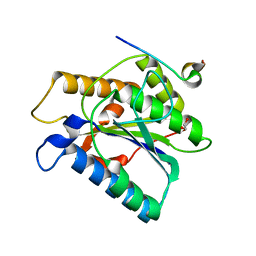 | |
1B67
 
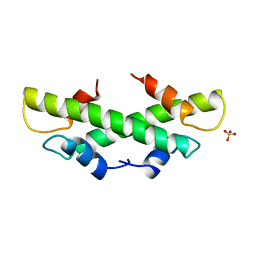 | | CRYSTAL STRUCTURE OF THE HISTONE HMFA FROM METHANOTHERMUS FERVIDUS | | 分子名称: | PROTEIN (HISTONE HMFA), SULFATE ION | | 著者 | Decanniere, K, Sandman, K, Reeve, J.N, Heinemann, U. | | 登録日 | 1999-01-19 | | 公開日 | 2000-01-17 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.48 Å) | | 主引用文献 | Crystal structures of recombinant histones HMfA and HMfB from the hyperthermophilic archaeon Methanothermus fervidus.
J.Mol.Biol., 303, 2000
|
|
1ATA
 
 | |
1G96
 
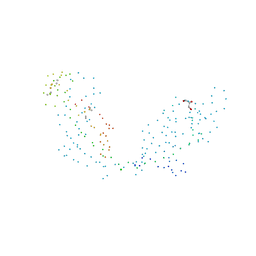 | | HUMAN CYSTATIN C; DIMERIC FORM WITH 3D DOMAIN SWAPPING | | 分子名称: | CHLORIDE ION, CYSTATIN C, GLYCEROL | | 著者 | Janowski, R, Kozak, M, Jankowska, E, Grzonka, Z, Grubb, A, Abrahamson, M, Jaskolski, M. | | 登録日 | 2000-11-22 | | 公開日 | 2001-04-06 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Human cystatin C, an amyloidogenic protein, dimerizes through three-dimensional domain swapping.
Nat.Struct.Biol., 8, 2001
|
|
5WOC
 
 | |
5WCV
 
 | |
1ATE
 
 | |
2AI6
 
 | |
5Z5W
 
 | |
6TI7
 
 | | Mixing Abeta(1-40) and Abeta(1-42) peptides generates unique amyloid fibrils | | 分子名称: | Amyloid-beta precursor protein | | 著者 | Cerofolini, L, Ravera, E, Bologna, S, Wiglenda, T, Boddrich, A, Purfurst, B, Benilova, A, Korsak, M, Gallo, G, Rizzo, D, Gonnelli, L, Fragai, M, De Strooper, B, Wanker, E.E, Luchinat, C. | | 登録日 | 2019-11-21 | | 公開日 | 2020-07-22 | | 最終更新日 | 2024-06-19 | | 実験手法 | SOLID-STATE NMR | | 主引用文献 | Mixing A beta (1-40) and A beta (1-42) peptides generates unique amyloid fibrils.
Chem.Commun.(Camb.), 56, 2020
|
|
1YHD
 
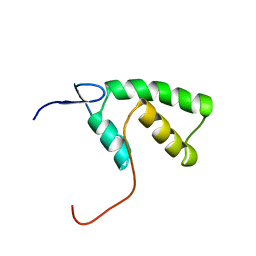 | | The solution structure of YGGX from Escherichia Coli | | 分子名称: | UPF0269 protein yggX | | 著者 | Osborne, M.J, Siddiqui, N, Landgraf, D, Pomposiello, P.J, Gehring, K. | | 登録日 | 2005-01-07 | | 公開日 | 2005-01-18 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The solution structure of the oxidative stress-related protein YggX from Escherichia coli.
Protein Sci., 14, 2005
|
|
1YG0
 
 | |
1GJS
 
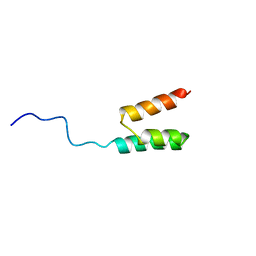 | | Solution structure of the Albumin binding domain of Streptococcal Protein G | | 分子名称: | IMMUNOGLOBULIN G BINDING PROTEIN G | | 著者 | Johansson, M.U, Frick, I.M, Nilsson, H, Kraulis, P.J, Hober, S, Jonasson, P, Nygren, A.P, Uhlen, M, Bjorck, L, Drakenberg, T, Forsen, S, Wikstrom, M. | | 登録日 | 2001-08-02 | | 公開日 | 2001-08-09 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure, Specificity, and Mode of Interaction for Bacterial Albumin-Binding Modules
J.Biol.Chem., 277, 2002
|
|
1Y6U
 
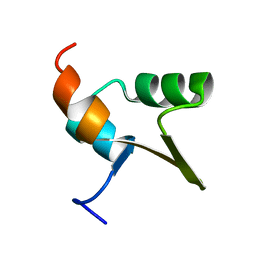 | |
5Z5R
 
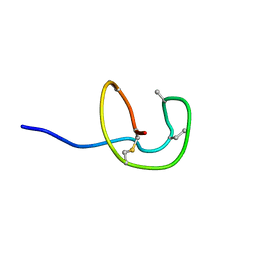 | | Nukacin ISK-1 in inactive state | | 分子名称: | Lantibiotic nukacin | | 著者 | Kohda, D, Fujinami, D. | | 登録日 | 2018-01-19 | | 公開日 | 2018-11-28 | | 最終更新日 | 2024-07-10 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The lantibiotic nukacin ISK-1 exists in an equilibrium between active and inactive lipid-II binding states.
Commun Biol, 1, 2018
|
|
1YMZ
 
 | | CC45, An Artificial WW Domain Designed Using Statistical Coupling Analysis | | 分子名称: | CC45 | | 著者 | Socolich, M, Lockless, S.W, Russ, W.P, Lee, H, Gardner, K.H, Ranganathan, R. | | 登録日 | 2005-01-22 | | 公開日 | 2005-09-27 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Evolutionary information for specifying a protein fold.
Nature, 437, 2005
|
|
1ATB
 
 | |
1ATD
 
 | |
