3VKI
 
 | |
4QCA
 
 | | Crystal structure of Vaccinia virus uracil-DNA glycosylase mutant R167AD4 | | 分子名称: | CHLORIDE ION, GLYCEROL, POTASSIUM ION, ... | | 著者 | Sartmatova, D, Nash, T, Schormann, N, Nuth, M, Ricciardi, R, Banerjee, S, Chattopadhyay, D. | | 登録日 | 2014-05-09 | | 公開日 | 2015-05-13 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystallization and preliminary X-ray diffraction analysis of three recombinant mutants of Vaccinia virus uracil DNA glycosylase.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
4Q16
 
 | |
4Q28
 
 | | Crystal Structure of the Plectin 1 and 2 Repeats of the Human Periplakin. Northeast Structural Genomics Consortium (NESG) Target HR9083A | | 分子名称: | Periplakin | | 著者 | Vorobiev, S, Lew, S, Seetharaman, J, Janjua, H, Xiao, R, O'Connell, P.T, Maglaqui, M, Wang, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2014-04-07 | | 公開日 | 2014-06-18 | | 実験手法 | X-RAY DIFFRACTION (2.64 Å) | | 主引用文献 | Crystal Structure of the Plectin 1 and 2 Repeats of Human Periplakin.
To be Published
|
|
4PWW
 
 | | Crystal Structure of Engineered Protein. Northeast Structural Genomics Consortium Target OR494. | | 分子名称: | ACETIC ACID, OR494, PHOSPHATE ION | | 著者 | Vorobiev, S, Lin, Y.-R, Seetharaman, J, Xiao, R, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2014-03-21 | | 公開日 | 2014-04-09 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.471 Å) | | 主引用文献 | Crystal Structure of Engineered Protein OR494.
To be Published
|
|
4Q29
 
 | | Ensemble Refinement of plu4264 protein from Photorhabdus luminescens | | 分子名称: | NICKEL (II) ION, SODIUM ION, plu4264 protein | | 著者 | Wang, F, Michalska, K, Li, H, Jedrzejczak, R, Babnigg, G, Bingman, C.A, Yennamalli, R, Weerth, S, Miller, M.D, Thomas, M.G, Joachimiak, A, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro), Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2014-04-07 | | 公開日 | 2014-05-07 | | 最終更新日 | 2015-02-11 | | 実験手法 | X-RAY DIFFRACTION (1.349 Å) | | 主引用文献 | Structure of a cupin protein Plu4264 from Photorhabdus luminescens subsp. laumondii TTO1 at 1.35 angstrom resolution.
Proteins, 83, 2015
|
|
4QC9
 
 | | Crystal structure of Vaccinia virus uracil-DNA glycosylase mutant 3GD4 | | 分子名称: | 1,2-ETHANEDIOL, SULFATE ION, Uracil-DNA glycosylase | | 著者 | Sartmatova, D, Nash, T, Schormann, N, Nuth, M, Ricciardi, R, Banerjee, S, Chattopadhyay, D. | | 登録日 | 2014-05-09 | | 公開日 | 2015-05-13 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.259 Å) | | 主引用文献 | Crystal structure of Vaccinia virus uracil-DNA glycosylase mutant 3GD4
To be Published
|
|
4QFB
 
 | | 1.99 A resolution structure of SeMet-CT263 (MTAN) from Chlamydia trachomatis | | 分子名称: | CT263 | | 著者 | Barta, M.L, Thomas, K, Lovell, S, Battaile, K.P, Schramm, V.L, Hefty, P.S. | | 登録日 | 2014-05-20 | | 公開日 | 2014-10-01 | | 最終更新日 | 2017-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.986 Å) | | 主引用文献 | Structural and Biochemical Characterization of Chlamydia trachomatis Hypothetical Protein CT263 Supports That Menaquinone Synthesis Occurs through the Futalosine Pathway.
J.Biol.Chem., 289, 2014
|
|
4QB9
 
 | | Crystal structure of Mycobacterium smegmatis Eis in complex with paromomycin | | 分子名称: | Enhanced intracellular survival protein, PAROMOMYCIN, SULFATE ION | | 著者 | Kim, K.H, Ahn, D.R, Yoon, H.J, Yang, J.K, Suh, S.W. | | 登録日 | 2014-05-06 | | 公開日 | 2015-04-01 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (3.293 Å) | | 主引用文献 | Structure of Mycobacterium smegmatis Eis in complex with paromomycin.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
4QA9
 
 | | Ensemble refinement of an epoxide hydrolase from Streptomyces carzinostaticus subsp. neocarzinostaticus. | | 分子名称: | 1,2-ETHANEDIOL, Epoxide hydrolase, SULFATE ION | | 著者 | Wang, F, Tan, K, Bigelow, L, Clancy, S, Babnigg, G, Bingman, C.A, Yennamalli, R, Lohman, J, Ma, M, Shen, B, Joachimiak, A, Phillips Jr, G.N, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2014-05-02 | | 公開日 | 2014-05-21 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.56 Å) | | 主引用文献 | Ensemble refinement of an epoxide hydrolase from Streptomyces carzinostaticus subsp. neocarzinostaticus.
To be Published
|
|
4Q9T
 
 | |
4QKX
 
 | | Structure of beta2 adrenoceptor bound to a covalent agonist and an engineered nanobody | | 分子名称: | 4-[(1R)-1-hydroxy-2-({2-[3-methoxy-4-(2-sulfanylethoxy)phenyl]ethyl}amino)ethyl]benzene-1,2-diol, Beta-2 adrenergic receptor, R9 protein, ... | | 著者 | Weichert, D, Kruse, A.C, Manglik, A, Hiller, C, Zhang, C, Huebner, H, Kobilka, B.K, Gmeiner, P. | | 登録日 | 2014-06-10 | | 公開日 | 2014-07-23 | | 最終更新日 | 2017-06-28 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Covalent agonists for studying G protein-coupled receptor activation.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4QQZ
 
 | | Crystal structure of T. fusca Cas3-AMPPNP | | 分子名称: | CRISPR-associated helicase, Cas3 family, DNA (5'-D(P*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3'), ... | | 著者 | Ke, A, Huo, Y, Nam, K.H. | | 登録日 | 2014-06-30 | | 公開日 | 2014-08-27 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.93 Å) | | 主引用文献 | Structures of CRISPR Cas3 offer mechanistic insights into Cascade-activated DNA unwinding and degradation.
Nat.Struct.Mol.Biol., 21, 2014
|
|
4QQY
 
 | | Crystal structure of T. fusca Cas3-ADP | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, CRISPR-associated helicase, Cas3 family, ... | | 著者 | Ke, A, Huo, Y, Nam, K.H. | | 登録日 | 2014-06-30 | | 公開日 | 2014-08-27 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (3.12 Å) | | 主引用文献 | Structures of CRISPR Cas3 offer mechanistic insights into Cascade-activated DNA unwinding and degradation.
Nat.Struct.Mol.Biol., 21, 2014
|
|
4QX0
 
 | | Cry3A Toxin structure obtained by Serial Femtosecond Crystallography from in vivo grown crystals isolated from Bacillus thuringiensis and data processed with the cctbx.xfel software suite | | 分子名称: | Pesticidal crystal protein cry3Aa | | 著者 | Sawaya, M.R, Cascio, D, Gingery, M, Rodriguez, J, Goldschmidt, L, Colletier, J.-P, Messerschmidt, M, Boutet, S, Koglin, J.E, Williams, G.J, Brewster, A.S, Nass, K, Hattne, J, Botha, S, Doak, R.B, Shoeman, R.L, DePonte, D.P, Park, H.-W, Federici, B.A, Sauter, N.K, Schlichting, I, Eisenberg, D. | | 登録日 | 2014-07-17 | | 公開日 | 2014-08-13 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Protein crystal structure obtained at 2.9 angstrom resolution from injecting bacterial cells into an X-ray free-electron laser beam.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4QT2
 
 | | Crystal Structure of the FK506-Binding Domain of Plasmodium Falciparum FKBP35 in complex with Rapamycin | | 分子名称: | FK506-binding protein (FKBP)-type peptidyl-propyl isomerase, IMIDAZOLE, RAPAMYCIN IMMUNOSUPPRESSANT DRUG | | 著者 | Bianchin, A, Allemand, F, Bell, A, Chubb, A.J, Guichou, J.-F. | | 登録日 | 2014-07-07 | | 公開日 | 2015-06-10 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.44 Å) | | 主引用文献 | Two crystal structures of the FK506-binding domain of Plasmodium falciparum FKBP35 in complex with rapamycin at high resolution
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4R80
 
 | | Crystal Structure of a De Novo Designed Beta Sheet Protein, Cystatin Fold, Northeast Structural Genomics Consortium (NESG) Target OR486 | | 分子名称: | OR486 | | 著者 | Guan, R, Marcos, E, O'Connell, P, Seetharaman, J, Janjua, H, Xiao, R, Maglaqui, M, Acton, T.B, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2014-08-29 | | 公開日 | 2014-09-24 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.445 Å) | | 主引用文献 | Crystal Structure of an engineered protein with denovo beta sheet design, Northeast Structural Genomics Consortium (NESG) Target OR486
To be Published
|
|
4R1F
 
 | |
4QX3
 
 | | Cry3A Toxin structure obtained by injecting Bacillus thuringiensis cells in an XFEL beam, collecting data by serial femtosecond crystallographic methods and processing data with the CrystFEL software suite | | 分子名称: | Pesticidal crystal protein cry3Aa | | 著者 | Sawaya, M.R, Cascio, D, Gingery, M, Rodriguez, J, Goldschmidt, L, Colletier, J.-P, Messerschmidt, M, Boutet, S, Koglin, J.E, Williams, G.J, Brewster, A.S, Nass, K, Hattne, J, Botha, S, Doak, R.B, Shoeman, R.L, DePonte, D.P, Park, H.-W, Federici, B.A, Sauter, N.K, Schlichting, I, Eisenberg, D. | | 登録日 | 2014-07-17 | | 公開日 | 2014-08-13 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Protein crystal structure obtained at 2.9 angstrom resolution from injecting bacterial cells into an X-ray free-electron laser beam.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4R31
 
 | |
4R4E
 
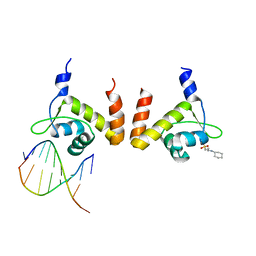 | | Structure of GlnR-DNA complex | | 分子名称: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, DNA (5'-D(*AP*TP*TP*CP*TP*GP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*CP*AP*GP*TP*A)-3'), ... | | 著者 | Schumacher, M.A. | | 登録日 | 2014-08-19 | | 公開日 | 2015-03-04 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.57 Å) | | 主引用文献 | Structures of regulatory machinery reveal novel molecular mechanisms controlling B. subtilis nitrogen homeostasis.
Genes Dev., 29, 2015
|
|
4QT3
 
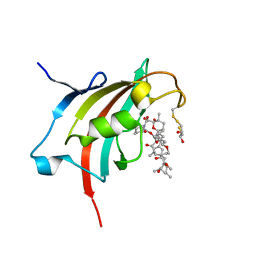 | | Crystal structure resolution of Plasmodium falciparum FK506 binding domain (FKBP35) in complex with Rapamycin at 1.4A resolution | | 分子名称: | (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, FK506-binding protein (FKBP)-type peptidyl-propyl isomerase, RAPAMYCIN IMMUNOSUPPRESSANT DRUG | | 著者 | Bianchin, A, Allemand, F, Bell, A, Chubb, A.J, Guichou, J.-F. | | 登録日 | 2014-07-07 | | 公開日 | 2015-06-10 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Two crystal structures of the FK506-binding domain of Plasmodium falciparum FKBP35 in complex with rapamycin at high resolution
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4QQX
 
 | | Crystal structure of T. fusca Cas3-ATP | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, CRISPR-associated helicase, Cas3 family, ... | | 著者 | Ke, A, Huo, Y, Nam, K.H. | | 登録日 | 2014-06-30 | | 公開日 | 2014-08-20 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.34 Å) | | 主引用文献 | Structures of CRISPR Cas3 offer mechanistic insights into Cascade-activated DNA unwinding and degradation.
Nat.Struct.Mol.Biol., 21, 2014
|
|
4QX2
 
 | | Cry3A Toxin structure obtained by injecting Bacillus thuringiensis cells in an XFEL beam, collecting data by serial femtosecond crystallographic methods and processing data with the cctbx.xfel software suite | | 分子名称: | Pesticidal crystal protein cry3Aa | | 著者 | Sawaya, M.R, Cascio, D, Gingery, M, Rodriguez, J, Goldschmidt, L, Colletier, J.-P, Messerschmidt, M, Boutet, S, Koglin, J.E, Williams, G.J, Brewster, A.S, Nass, K, Hattne, J, Botha, S, Doak, R.B, Shoeman, R.L, DePonte, D.P, Park, H.-W, Federici, B.A, Sauter, N.K, Schlichting, I, Eisenberg, D. | | 登録日 | 2014-07-17 | | 公開日 | 2014-08-13 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Protein crystal structure obtained at 2.9 angstrom resolution from injecting bacterial cells into an X-ray free-electron laser beam.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4QX6
 
 | | CRYSTAL STRUCTURE OF GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE FROM STREPTOCOCCUS AGALACTIAE NEM316 at 2.46 ANGSTROM RESOLUTION | | 分子名称: | 1,2-ETHANEDIOL, Glyceraldehyde 3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Ayres, C.A, Schormann, N, Banerjee, S, Chattopadhyay, D. | | 登録日 | 2014-07-18 | | 公開日 | 2014-10-15 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.46 Å) | | 主引用文献 | Structure of Streptococcus agalactiae glyceraldehyde-3-phosphate dehydrogenase holoenzyme reveals a novel surface.
Acta Crystallogr F Struct Biol Commun, 70, 2014
|
|
