5PTD
 
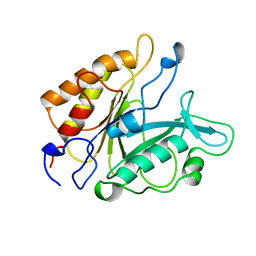 | | PHOSPHATIDYLINOSITOL-SPECIFIC PHOSPHOLIPASE C MUTANT H32A | | 分子名称: | PHOSPHATIDYLINOSITOL-SPECIFIC PHOSPHOLIPASE C | | 著者 | Heinz, D.W. | | 登録日 | 1997-07-18 | | 公開日 | 1998-01-21 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Probing the roles of active site residues in phosphatidylinositol-specific phospholipase C from Bacillus cereus by site-directed mutagenesis.
Biochemistry, 36, 1997
|
|
4BFF
 
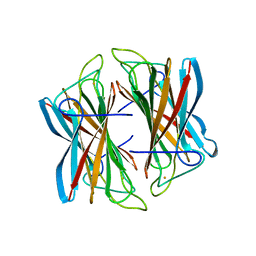 | | Superoxide reductase (Neelaredoxin) from Archaeoglobus fulgidus in the reduced form | | 分子名称: | FE (II) ION, SUPEROXIDE REDUCTASE | | 著者 | Bandeiras, T.M, Rodrigues, J.V, Sousa, C.M, Barradas, A.R, Pinho, F.G, Pinto, A.F, Teixeira, M, Matias, P.M, Romao, C.V. | | 登録日 | 2013-03-18 | | 公開日 | 2014-03-26 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Understanding the Role of Key Residues in the Superoxide Reductase Molecular Mechanism, Exploring Archaeoglobus Fulgidus Sor Structure
To be Published
|
|
8HZ4
 
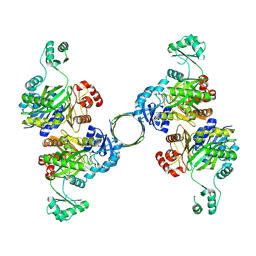 | |
8INL
 
 | | LSD1 in complex with S2172 | | 分子名称: | 3-[3,5-bis(fluoranyl)-2-phenylmethoxy-phenyl]propanal, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | 著者 | Niwa, H, Sato, S, Umehara, T. | | 登録日 | 2023-03-10 | | 公開日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.62 Å) | | 主引用文献 | LSD1 in complex with S2172
To Be Published
|
|
5QAK
 
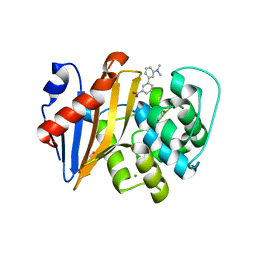 | | OXA-48 IN COMPLEX WITH COMPOUND 14 | | 分子名称: | 1,2-ETHANEDIOL, 3-[3-(dimethylamino)phenyl]benzoic acid, Beta-lactamase, ... | | 著者 | Lund, B.A, Leiros, H.K.S. | | 登録日 | 2017-07-11 | | 公開日 | 2018-01-10 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | A focused fragment library targeting the antibiotic resistance enzyme - Oxacillinase-48: Synthesis, structural evaluation and inhibitor design.
Eur J Med Chem, 145, 2018
|
|
4ASI
 
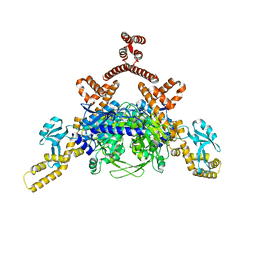 | | Crystal structure of human ACACA C-terminal domain | | 分子名称: | ACETYL-COA CARBOXYLASE 1 | | 著者 | Froese, D.S, Muniz, J.R.C, Kiyani, W, Krojer, T, Vollmar, M, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Oppermann, U, Yue, W.W. | | 登録日 | 2012-05-01 | | 公開日 | 2012-08-29 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal Structure of Human Acaca C-Terminal Domain
To be Published
|
|
7C8Z
 
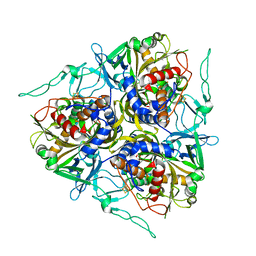 | |
4ASU
 
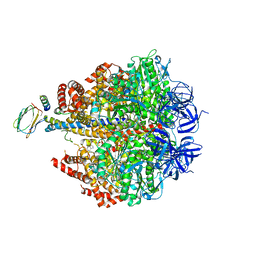 | | F1-ATPase in which all three catalytic sites contain bound nucleotide, with magnesium ion released in the Empty site | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ATP SYNTHASE SUBUNIT ALPHA, MITOCHONDRIAL, ... | | 著者 | Rees, D.M, Montgomery, M.G, Leslie, A.G.W, Walker, J.E. | | 登録日 | 2012-05-03 | | 公開日 | 2012-06-27 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural Evidence of a New Catalytic Intermediate in the Pathway of ATP Hydrolysis by F1-ATPase from Bovine Heart Mitochondria.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5QNN
 
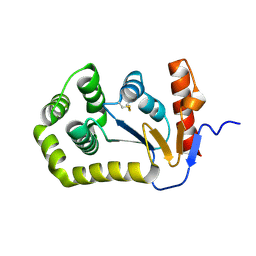 | |
8IXZ
 
 | |
4CAR
 
 | | Structure of bovine endothelial nitric oxide synthase heme domain in complex with 7-((3-Fluorophenethylamino)methyl)quinolin-2-amine | | 分子名称: | 5,6,7,8-TETRAHYDROBIOPTERIN, 7-[[2-(3-fluorophenyl)ethylamino]methyl]quinolin-2-amine, ACETATE ION, ... | | 著者 | Li, H, Poulos, T.L. | | 登録日 | 2013-10-08 | | 公開日 | 2014-02-19 | | 最終更新日 | 2014-03-12 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Simplified 2-Aminoquinoline-Based Scaffold for Potent and Selective Neuronal Nitric Oxide Synthase Inhibition.
J.Med.Chem., 57, 2014
|
|
4CCL
 
 | | X-Ray structure of E. coli ycfD | | 分子名称: | 50S RIBOSOMAL PROTEIN L16 ARGININE HYDROXYLASE, MANGANESE (II) ION, SULFATE ION | | 著者 | McDonough, M.A, Ho, C.H, Kershaw, N.J, Schofield, C.J. | | 登録日 | 2013-10-23 | | 公開日 | 2014-04-30 | | 最終更新日 | 2020-06-03 | | 実験手法 | X-RAY DIFFRACTION (2.596 Å) | | 主引用文献 | Ribosomal oxygenases are structurally conserved from prokaryotes to humans.
Nature, 510, 2014
|
|
5QYY
 
 | | PanDDA analysis group deposition -- Auto-refined data of Aar2/RNaseH for ground state model 14 | | 分子名称: | A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | 著者 | Weiss, M.S, Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G. | | 登録日 | 2020-02-12 | | 公開日 | 2020-06-10 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.71 Å) | | 主引用文献 | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
7CUQ
 
 | | 2.55-Angstrom Cryo-EM structure of Cytochrome bo3 from Escherichia coli in Native Membrane | | 分子名称: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, COPPER (II) ION, Cytochrome bo(3) ubiquinol oxidase subunit 1, ... | | 著者 | Li, J, Han, L, Gennis, R.B, Zhu, J.P, Zhang, K. | | 登録日 | 2020-08-24 | | 公開日 | 2021-08-25 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (2.64 Å) | | 主引用文献 | Cryo-EM structures of Escherichia coli cytochrome bo3 reveal bound phospholipids and ubiquinone-8 in a dynamic substrate binding site.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CUW
 
 | | Ubiquinol Binding Site of Cytochrome bo3 from Escherichia coli | | 分子名称: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, COPPER (II) ION, Cytochrome bo(3) ubiquinol oxidase subunit 1, ... | | 著者 | Li, J, Han, L, Gennis, R.B, Zhu, J.P, Zhang, K. | | 登録日 | 2020-08-25 | | 公開日 | 2021-08-25 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (2.63 Å) | | 主引用文献 | Cryo-EM structures of Escherichia coli cytochrome bo3 reveal bound phospholipids and ubiquinone-8 in a dynamic substrate binding site.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CUB
 
 | | 2.55-Angstrom Cryo-EM structure of Cytochrome bo3 from Escherichia coli in Native Membrane | | 分子名称: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, COPPER (II) ION, Cytochrome bo(3) ubiquinol oxidase subunit 1, ... | | 著者 | Li, J, Han, L, Gennis, R.B, Zhu, J.P, Zhang, K. | | 登録日 | 2020-08-22 | | 公開日 | 2021-08-25 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (2.55 Å) | | 主引用文献 | Cryo-EM structures of Escherichia coli cytochrome bo3 reveal bound phospholipids and ubiquinone-8 in a dynamic substrate binding site.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
4AD7
 
 | | Crystal structure of full-length N-glycosylated human glypican-1 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYPICAN-1 | | 著者 | Svensson, G, Awad, W, Mani, K, Logan, D.T. | | 登録日 | 2011-12-22 | | 公開日 | 2012-03-21 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.945 Å) | | 主引用文献 | Crystal Structure of N-Glycosylated Human Glypican-1 Core Protein: Structure of Two Loops Evolutionarily Conserved in Vertebrate Glypican-1.
J.Biol.Chem., 287, 2012
|
|
7D0Q
 
 | |
5R2Q
 
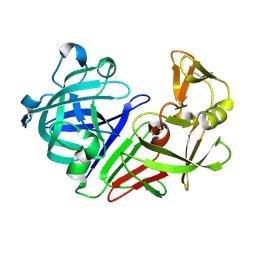 | | PanDDA analysis group deposition -- Auto-refined data of Endothiapepsin for ground state model 14, DMSO-Free | | 分子名称: | Endothiapepsin | | 著者 | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | 登録日 | 2020-02-13 | | 公開日 | 2020-06-03 | | 最終更新日 | 2020-07-08 | | 実験手法 | X-RAY DIFFRACTION (1.038 Å) | | 主引用文献 | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
3ZV8
 
 | |
3ZVF
 
 | | 3C protease of Enterovirus 68 complexed with Michael receptor inhibitor 85 | | 分子名称: | 3C PROTEASE, N-[(benzyloxy)carbonyl]-O-tert-butyl-L-seryl-N-{(2R)-5-ethoxy-5-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]pentan-2-yl}-L-phenylalaninamide | | 著者 | Tan, J, Perbandt, M, Mesters, J.R, Hilgenfeld, R. | | 登録日 | 2011-07-24 | | 公開日 | 2012-08-08 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | 3C Protease of Enterovirus 68: Structure-Based Design of Michael Acceptor Inhibitors and Their Broad-Spectrum Antiviral Effects Against Picornaviruses.
J.Virol., 87, 2013
|
|
3ZV9
 
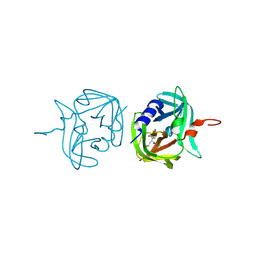 | | 3C protease of Enterovirus 68 complexed with Michael receptor inhibitor 74 | | 分子名称: | 3C PROTEASE, ETHYL (4R)-4-[(TERT-BUTOXYCARBONYL)AMINO]-5-[(3S)-2-OXOPYRROLIDIN-3-YL]PENTANOATE | | 著者 | Tan, J, Perbandt, M, Mesters, J.R, Hilgenfeld, R. | | 登録日 | 2011-07-24 | | 公開日 | 2012-08-08 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | 3C Protease of Enterovirus 68: Structure-Based Design of Michael Acceptor Inhibitors and Their Broad-Spectrum Antiviral Effects Against Picornaviruses.
J.Virol., 87, 2013
|
|
3ZVE
 
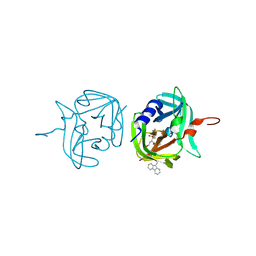 | | 3C protease of Enterovirus 68 complexed with Michael receptor inhibitor 84 | | 分子名称: | 3C PROTEASE, O-tert-butyl-N-[(9H-fluoren-9-ylmethoxy)carbonyl]-L-threonyl-N-{(2R)-5-ethoxy-5-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]pentan-2-yl}-L-phenylalaninamide | | 著者 | Tan, J, Perbandt, M, Mesters, J.R, Hilgenfeld, R. | | 登録日 | 2011-07-24 | | 公開日 | 2012-08-08 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | 3C Protease of Enterovirus 68: Structure-Based Design of Michael Acceptor Inhibitors and Their Broad-Spectrum Antiviral Effects Against Picornaviruses.
J.Virol., 87, 2013
|
|
4CAM
 
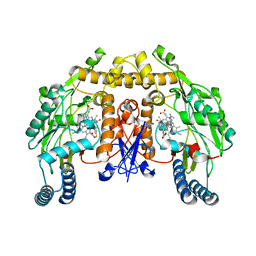 | | Structure of rat neuronal nitric oxide synthase heme domain in complex with 7-((3-Fluorophenethylamino)methyl)quinolin-2-amine | | 分子名称: | 5,6,7,8-TETRAHYDROBIOPTERIN, 7-[[2-(3-fluorophenyl)ethylamino]methyl]quinolin-2-amine, ACETATE ION, ... | | 著者 | Li, H, Poulos, T.L. | | 登録日 | 2013-10-08 | | 公開日 | 2014-02-19 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.83 Å) | | 主引用文献 | Simplified 2-Aminoquinoline-Based Scaffold for Potent and Selective Neuronal Nitric Oxide Synthase Inhibition.
J.Med.Chem., 57, 2014
|
|
7DA9
 
 | |
