3HP8
 
 | | Crystal structure of a designed Cyanovirin-N homolog lectin; LKAMG, bound to sucrose | | 分子名称: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Cyanovirin-N-like protein, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | 著者 | Koharudin, L.M.I, Furey, W, Gronenborn, A.M. | | 登録日 | 2009-06-03 | | 公開日 | 2009-06-23 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | A designed chimeric cyanovirin-N homolog lectin: Structure and molecular basis of sucrose binding.
Proteins, 77, 2009
|
|
6JXU
 
 | | SUMO1 bound to SLS4-SIM peptide from ICP0 | | 分子名称: | Small ubiquitin-related modifier, viral protein | | 著者 | Hembram, D.S.S, Negi, H, Shet, D, Das, R. | | 登録日 | 2019-04-25 | | 公開日 | 2020-02-05 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The Viral SUMO-Targeted Ubiquitin Ligase ICP0 is Phosphorylated and Activated by Host Kinase Chk2.
J.Mol.Biol., 432, 2020
|
|
6JXW
 
 | | Complex of SUMO2 bound SLS4 from ICP0. | | 分子名称: | SLS4-SIM from Ubiquitin E3 ligase ICP0, Small ubiquitin-related modifier 2 | | 著者 | Hembram, D.S.S, Negi, H, Shet, D, Das, R. | | 登録日 | 2019-04-25 | | 公開日 | 2020-02-05 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The Viral SUMO-Targeted Ubiquitin Ligase ICP0 is Phosphorylated and Activated by Host Kinase Chk2.
J.Mol.Biol., 432, 2020
|
|
6SDY
 
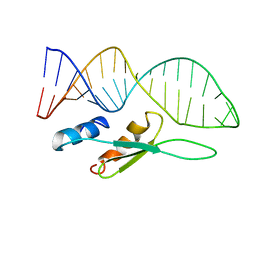 | |
6SDW
 
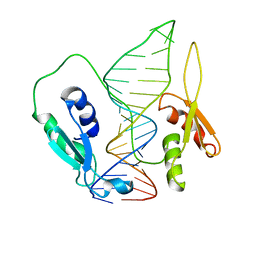 | |
5DO6
 
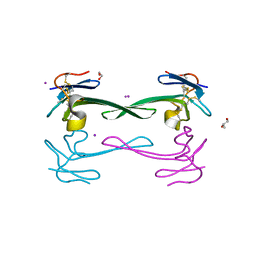 | | Crystal structure of Dendroaspis polylepis venom mambalgin-1 T23A mutant | | 分子名称: | 1,2-ETHANEDIOL, IODIDE ION, Mambalgin-1, ... | | 著者 | Stura, E.A, Tepshi, L, Kessler, P, Gilles, M, Servent, D. | | 登録日 | 2015-09-10 | | 公開日 | 2015-12-30 | | 最終更新日 | 2017-01-25 | | 実験手法 | X-RAY DIFFRACTION (1.697 Å) | | 主引用文献 | Mambalgin-1 Pain-relieving Peptide, Stepwise Solid-phase Synthesis, Crystal Structure, and Functional Domain for Acid-sensing Ion Channel 1a Inhibition.
J.Biol.Chem., 291, 2016
|
|
5OEO
 
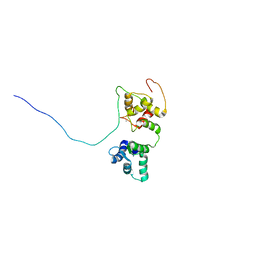 | | Solution structure of the complex of TRPV5(655-725) with a Calmodulin E32Q/E68Q double mutant | | 分子名称: | CALCIUM ION, Calmodulin-1, Transient receptor potential cation channel subfamily V member 5 | | 著者 | Vuister, G.W, Bokhovchuk, F.M, Bate, N, Kovalevskaya, N, Goult, B.T, Spronk, C.A.E.M. | | 登録日 | 2017-07-09 | | 公開日 | 2018-04-25 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The Structural Basis of Calcium-Dependent Inactivation of the Transient Receptor Potential Vanilloid 5 Channel.
Biochemistry, 57, 2018
|
|
1VEG
 
 | | Solution Structure of RSGI RUH-012, a UBA Domain from Mouse cDNA | | 分子名称: | NEDD8 ultimate buster-1 | | 著者 | Abe, T, Hirota, H, Izumi, K, Yoshida, M, Yamazaki, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2004-03-31 | | 公開日 | 2004-09-30 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution Structure of RSGI RUH-012, a UBA Domain from Mouse cDNA
To be Published
|
|
5KES
 
 | | Solution structure of the yeast Ddi1 HDD domain | | 分子名称: | DNA damage-inducible protein 1 | | 著者 | Trempe, J.-F, Ratcliffe, C, Veverka, V, Saskova, K, Gehring, K. | | 登録日 | 2016-06-10 | | 公開日 | 2016-10-05 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural studies of the yeast DNA damage-inducible protein Ddi1 reveal domain architecture of this eukaryotic protein family.
Sci Rep, 6, 2016
|
|
5JPW
 
 | | Molecular basis for protein recognition specificity of the DYNLT1/Tctex1 canonical binding groove. Characterization of the interaction with activin receptor IIB | | 分子名称: | Dynein light chain Tctex-type 1,Cytoplasmic dynein 1 intermediate chain 2 | | 著者 | Rodriguez-Crespo, I, Merino-Gracia, J, Bruix, M, Zamora-Carreras, H. | | 登録日 | 2016-05-04 | | 公開日 | 2016-08-17 | | 最終更新日 | 2024-07-03 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Molecular Basis for the Protein Recognition Specificity of the Dynein Light Chain DYNLT1/Tctex1: CHARACTERIZATION OF THE INTERACTION WITH ACTIVIN RECEPTOR IIB.
J.Biol.Chem., 291, 2016
|
|
8JB4
 
 | | lipopolysaccharide-binding domain-LBDB | | 分子名称: | Antilipopolysaccharide factor D | | 著者 | Huang, J, Qin, Z. | | 登録日 | 2023-05-08 | | 公開日 | 2024-03-13 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Machine learning and genetic algorithm-guided directed evolution for the development of small-molecule antibiotics originating from antimicrobial peptides
To Be Published
|
|
4GS9
 
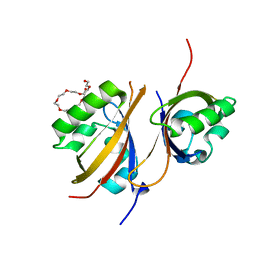 | | Crystal structure of the high affinity heterodimer of HIF2 alpha and ARNT C-terminal PAS domains in complex with an inactive benzoxadiazole antagonist | | 分子名称: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain-containing protein 1, ... | | 著者 | Scheuermann, T.H, Gardner, K.H. | | 登録日 | 2012-08-27 | | 公開日 | 2013-04-03 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.72 Å) | | 主引用文献 | Development of Inhibitors of the PAS-B Domain of the HIF-2 alpha Transcription Factor
J.Med.Chem., 56, 2013
|
|
6PV0
 
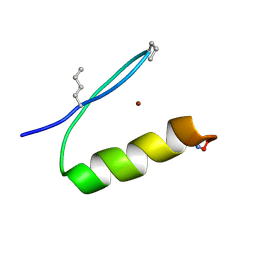 | |
7Y7L
 
 | | Solution structure of zinc finger domain 2 of human ZFAND1 | | 分子名称: | AN1-type zinc finger protein 1, ZINC ION | | 著者 | Fang, P.J, Lai, C.H, Ko, K.T, Chang, C.F, Hsu, S.T.D. | | 登録日 | 2022-06-22 | | 公開日 | 2023-06-28 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural basis of p97 recognition by human ZFAND1
To Be Published
|
|
7YAB
 
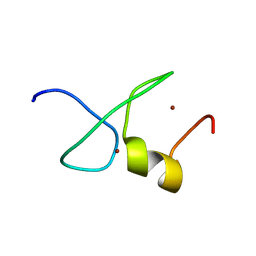 | | Solution structure of zinc finger domain 1 of human ZFAND1 | | 分子名称: | AN1-type zinc finger protein 1, ZINC ION | | 著者 | Fang, P.J, Lai, C.H, Ko, K.T, Chang, C.F, Hsu, S.T.D. | | 登録日 | 2022-06-27 | | 公開日 | 2023-06-28 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural basis of p97 recognition by human ZFAND1
To Be Published
|
|
7Y0I
 
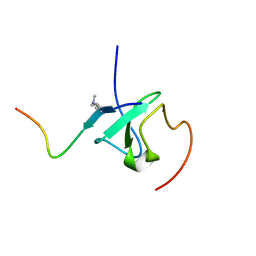 | |
8IS3
 
 | | Structural model for the micelle-bound indolicidin-like peptide in solution | | 分子名称: | Indolicidin-like antimicrobial peptide | | 著者 | Kim, B, Ko, Y.H, Kim, J, Lee, J, Nam, C.H, Kim, J.H. | | 登録日 | 2023-03-20 | | 公開日 | 2024-03-20 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural model for the micelle-bound indolicidin-like peptide in solution
To Be Published
|
|
5TX8
 
 | |
8BXJ
 
 | |
7LGL
 
 | |
7LI2
 
 | |
7LXC
 
 | |
7LIF
 
 | |
7LC8
 
 | | SARS-CoV-2 spike Protein TM domain | | 分子名称: | Spike protein S2' | | 著者 | Fu, Q, Chou, J.J. | | 登録日 | 2021-01-10 | | 公開日 | 2021-06-23 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | A Trimeric Hydrophobic Zipper Mediates the Intramembrane Assembly of SARS-CoV-2 Spike.
J.Am.Chem.Soc., 143, 2021
|
|
8C2Q
 
 | | Silver ion-bound structure of the silver specific chaperone SilF needed for bacterial silver resistance | | 分子名称: | Copper ABC transporter substrate-binding protein, SILVER ION | | 著者 | Monneau, Y.R, Walker, O, Hologne, M. | | 登録日 | 2022-12-22 | | 公開日 | 2023-10-25 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The battle for silver binding: How the interplay between the SilE, SilF, and SilB proteins contributes to the silver efflux pump mechanism.
J.Biol.Chem., 299, 2023
|
|
