3CZE
 
 | |
3HGN
 
 | | Structure of porcine pancreatic elastase complexed with a potent peptidyl inhibitor FR130180 determined by neutron crystallography | | 分子名称: | 4-[[(2S)-3-methyl-1-oxo-1-[(2S)-2-[[(3S)-1,1,1-trifluoro-4-methyl-2-oxo-pentan-3-yl]carbamoyl]pyrrolidin-1-yl]butan-2-yl]carbamoyl]benzoic acid, CALCIUM ION, Elastase-1, ... | | 著者 | Tamada, T, Kinoshita, T, Kuroki, R, Tada, T. | | 登録日 | 2009-05-14 | | 公開日 | 2009-07-28 | | 最終更新日 | 2023-11-01 | | 実験手法 | NEUTRON DIFFRACTION (1.65 Å), X-RAY DIFFRACTION | | 主引用文献 | Combined High-Resolution Neutron and X-ray Analysis of Inhibited Elastase Confirms the Active-Site Oxyanion Hole but Rules against a Low-Barrier Hydrogen Bond
J.Am.Chem.Soc., 131, 2009
|
|
5Q1I
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | 分子名称: | 3-(2-chlorophenyl)-N-[(1R)-1-(naphthalen-2-yl)ethyl]-5-(propan-2-yl)-1,2-oxazole-4-carboxamide, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | 著者 | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-05-31 | | 公開日 | 2017-07-05 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5FB8
 
 | | Structure of Interleukin-16 bound to the 14.1 antibody | | 分子名称: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Anti-IL-16 antibody 14.1 Fab domain Heavy Chain, ... | | 著者 | Hall, G, Cowan, R, Bayliss, R, Carr, M. | | 登録日 | 2015-12-14 | | 公開日 | 2016-06-08 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.07 Å) | | 主引用文献 | Structure of a Potential Therapeutic Antibody Bound to Interleukin-16 (IL-16): MECHANISTIC INSIGHTS AND NEW THERAPEUTIC OPPORTUNITIES.
J.Biol.Chem., 291, 2016
|
|
7XEY
 
 | | EDS1-PAD4 complexed with pRib-ADP | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5-O-phosphono-beta-D-ribofuranose, ADENOSINE-5'-DIPHOSPHATE, ... | | 著者 | Huang, S, Jia, A, Xiao, Y. | | 登録日 | 2022-03-31 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.29 Å) | | 主引用文献 | Identification and receptor mechanism of TIR-catalyzed small molecules in plant immunity.
Science, 377, 2022
|
|
4M14
 
 | |
5FMC
 
 | | Structure of D80A-fructofuranosidase from Xanthophyllomyces dendrorhous complexed with fructose and BIS-TRIS propane buffer | | 分子名称: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Ramirez-Escudero, M, Sanz-Aparicio, J. | | 登録日 | 2015-11-02 | | 公開日 | 2016-02-10 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.84 Å) | | 主引用文献 | Structural Analysis of Beta-Fructofuranosidase from Xanthophyllomyces Dendrorhous Reveals Unique Features and the Crucial Role of N-Glycosylation in Oligomerization and Activity
J.Biol.Chem., 291, 2016
|
|
4M15
 
 | |
5UWA
 
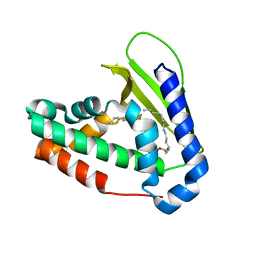 | |
5DJE
 
 | | Crystal structure of the zuotin homology domain (ZHD) from yeast Zuo1 | | 分子名称: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Shrestha, O.K, Bingman, C.A, Craig, E.A. | | 登録日 | 2015-09-02 | | 公開日 | 2016-09-28 | | 最終更新日 | 2019-12-25 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Dual interaction of the Hsp70 J-protein cochaperone Zuotin with the 40S and 60S ribosomal subunits.
Nat.Struct.Mol.Biol., 23, 2016
|
|
3BAW
 
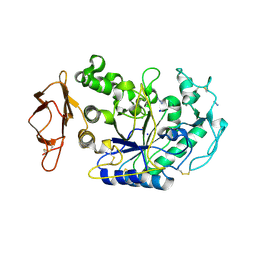 | |
4CBZ
 
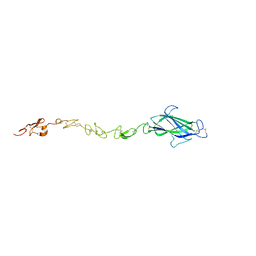 | | Notch ligand, Jagged-1, contains an N-terminal C2 domain | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEIN JAGGED-1, alpha-L-fucopyranose | | 著者 | Chilakuri, C.R, Sheppard, D, Ilagan, M.X.G, Holt, L.R, Abbott, F, Liang, S, Kopan, R, Handford, P.A, Lea, S.M. | | 登録日 | 2013-10-17 | | 公開日 | 2013-11-27 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural Analysis Uncovers Lipid-Binding Properties of Notch Ligands
Cell Rep., 5, 2013
|
|
4DNI
 
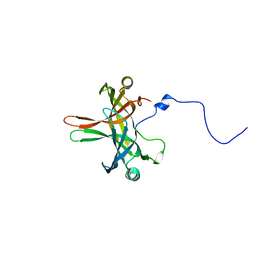 | | Structure of Editosome protein | | 分子名称: | Fusion protein of RNA-editing complex proteins MP42 and MP18 | | 著者 | Park, Y.-J, Hol, W. | | 登録日 | 2012-02-08 | | 公開日 | 2012-12-26 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Explorations of linked editosome domains leading to the discovery of motifs defining conserved pockets in editosome OB-folds.
J.Struct.Biol., 180, 2012
|
|
4D7R
 
 | | Crystal structure of a chimeric protein with the Sec7 domain of Rickettsia prowazekii RalF and the capping domain of Legionella pneumophila RalF | | 分子名称: | PROLINE/BETAINE TRANSPORTER, RALF | | 著者 | Folly-Klan, M, Sancerne, B, Alix, E, Roy, C.R, Cherfils, J, Campanacci, V. | | 登録日 | 2014-11-27 | | 公開日 | 2015-01-14 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | On the Use of Legionella/Rickettsia Chimeras to Investigate the Structure and Regulation of Rickettsia Effector Ralf.
J.Struct.Biol., 189, 2015
|
|
7Y07
 
 | | Crystal structure of Ricin A chain bound with (S)-2-amino-N-(1-hydroxy-3-phenylpropan-2-yl)-4-oxo-3,4-dihydropteridine-7-carboxamide | | 分子名称: | 2-azanyl-4-oxidanylidene-N-[(2S)-1-oxidanyl-3-phenyl-propan-2-yl]-3H-pteridine-7-carboxamide, Ricin A chain, SULFATE ION | | 著者 | Goto, M, Higashi, S, Ohba, T, Kawata, R, Nagatsu, K, Suzuki, S, Saito, R. | | 登録日 | 2022-06-03 | | 公開日 | 2022-12-21 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Conformational change in ricin toxin A-Chain: A critical factor for inhibitor binding to the secondary pocket.
Biochem.Biophys.Res.Commun., 627, 2022
|
|
5Q0T
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | 分子名称: | 2-phenyl-N-(propan-2-yl)-6-[(thiophen-2-yl)sulfonyl]-4,5,6,7-tetrahydro-1H-pyrrolo[2,3-c]pyridine-1-carboxamide, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | 著者 | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-05-31 | | 公開日 | 2017-07-05 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.14 Å) | | 主引用文献 | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
7YCF
 
 | | HYDROXYNITRILE LYASE FROM THE MILLIPEDE, Oxidus gracilis IN ACETONITRILE | | 分子名称: | 2-HYDROXY-2-METHYLPROPANENITRILE, CHLORIDE ION, Hydroxynitrile lyase, ... | | 著者 | Chaikaew, S, Watanabe, Y, Zheng, D, Motojima, F, Asano, Y. | | 登録日 | 2022-07-01 | | 公開日 | 2024-01-24 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | Structure-Based Site-Directed Mutagenesis of Hydroxynitrile Lyase from Cyanogenic Millipede, Oxidus gracilis for Hydrocyanation and Henry Reactions.
Chembiochem, 25, 2024
|
|
3ZNQ
 
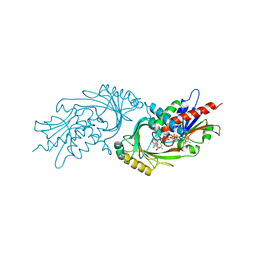 | | IN VITRO AND IN VIVO INHIBITION OF HUMAN D-AMINO ACID OXIDASE: REGULATION OF D-SERINE CONCENTRATION IN THE BRAIN | | 分子名称: | 3-PHENETHYL-4H-FURO[3,2-B]PYRROLE-5-CARBOXYLIC ACID, D-AMINO-ACID OXIDASE, FLAVIN-ADENINE DINUCLEOTIDE | | 著者 | Hopkins, S.C, Heffernan, M.L.R, Saraswat, L.D, Bowen, C.A, Melnick, L, Hardy, L.W, Orsini, M.A, Allen, M.S, Koch, P, Spear, K.L, Foglesong, R.J, Soukri, M, Chytil, M, Fang, Q.K, Jones, S.W, Varney, M.A, Panatier, A, Oliet, S.H.R, Pollegioni, L, Piubelli, L, Molla, G, Nardini, M, Large, T.H. | | 登録日 | 2013-02-15 | | 公開日 | 2013-05-15 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Structural, Kinetic, and Pharmacodynamic Mechanisms of D-Amino Acid Oxidase Inhibition by Small Molecules.
J.Med.Chem., 56, 2013
|
|
4CC1
 
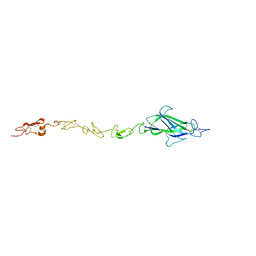 | | Notch ligand, Jagged-1, contains an N-terminal C2 domain | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | 著者 | Chilakuri, C.R, Sheppard, D, Ilagan, M.X.G, Holt, L.R, Abbott, F, Liang, S, Kopan, R, Handford, P.A, Lea, S.M. | | 登録日 | 2013-10-17 | | 公開日 | 2013-11-27 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.84 Å) | | 主引用文献 | Structural Analysis Uncovers Lipid-Binding Properties of Notch Ligands
Cell Rep., 5, 2013
|
|
4CC0
 
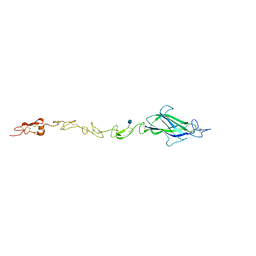 | | Notch ligand, Jagged-1, contains an N-terminal C2 domain | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, PROTEIN JAGGED-1, ... | | 著者 | Chilakuri, C.R, Sheppard, D, Ilagan, M.X.G, Holt, L.R, Abbott, F, Liang, S, Kopan, R, Handford, P.A, Lea, S.M. | | 登録日 | 2013-10-17 | | 公開日 | 2013-11-27 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.32 Å) | | 主引用文献 | Structural Analysis Uncovers Lipid-Binding Properties of Notch Ligands
Cell Rep., 5, 2013
|
|
4LX4
 
 | | Crystal Structure Determination of Pseudomonas stutzeri endoglucanase Cel5A using a Twinned Data Set | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Endoglucanase(Endo-1,4-beta-glucanase)protein | | 著者 | Dutoit, R, Delsaute, M, Berlemont, R, Van Elder, D, Galleni, M, Bauvois, C. | | 登録日 | 2013-07-29 | | 公開日 | 2014-07-30 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.556 Å) | | 主引用文献 | Crystal structure determination of Pseudomonas stutzeri A1501 endoglucanase Cel5A: the search for a molecular basis for glycosynthesis in GH5_5 enzymes.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
4LYP
 
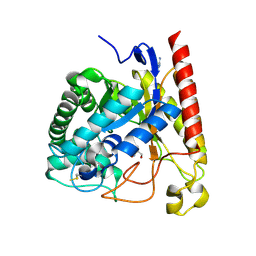 | | Crystal Structure of Glycoside Hydrolase Family 5 Mannosidase from Rhizomucor miehei | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Exo-beta-1,4-mannosidase, GUANIDINE | | 著者 | Jiang, Z.Q, Zhou, P, Yang, S.Q, Liu, Y, Yan, Q.J. | | 登録日 | 2013-07-31 | | 公開日 | 2014-08-06 | | 最終更新日 | 2014-11-26 | | 実験手法 | X-RAY DIFFRACTION (1.28 Å) | | 主引用文献 | Structural insights into the substrate specificity and transglycosylation activity of a fungal glycoside hydrolase family 5 beta-mannosidase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4L2N
 
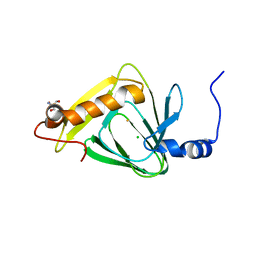 | | Understanding Extradiol Dioxygenase Mechanism in NAD+ Biosynthesis by Viewing Catalytic Intermediates - ligand-free structure | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-hydroxyanthranilate 3,4-dioxygenase, CHLORIDE ION, ... | | 著者 | Liu, F, Liu, A. | | 登録日 | 2013-06-04 | | 公開日 | 2015-05-06 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.74 Å) | | 主引用文献 | An Iron Reservoir to the Catalytic Metal: THE RUBREDOXIN IRON IN AN EXTRADIOL DIOXYGENASE.
J.Biol.Chem., 290, 2015
|
|
4LGL
 
 | | Crystal Structure of Glycine Decarboxylase P-protein from Synechocystis sp. PCC 6803, apo form | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, Glycine dehydrogenase [decarboxylating] | | 著者 | Hasse, D, Andersson, E, Carlsson, G, Masloboy, A, Hagemann, M, Bauwe, H, Andersson, I. | | 登録日 | 2013-06-28 | | 公開日 | 2013-10-16 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.0004 Å) | | 主引用文献 | Structure of the Homodimeric Glycine Decarboxylase P-protein from Synechocystis sp. PCC 6803 Suggests a Mechanism for Redox Regulation.
J.Biol.Chem., 288, 2013
|
|
4LLC
 
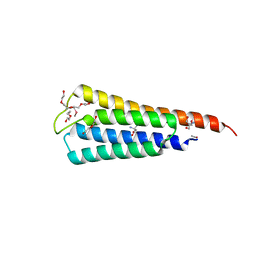 | | The crystal structure of R60E mutant of the histidine kinase (KinB) sensor domain from Pseudomonas aeruginosa PA01 | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, Probable two-component sensor, ... | | 著者 | Tan, K, Chhor, G, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2013-07-09 | | 公開日 | 2013-08-07 | | 最終更新日 | 2013-08-14 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The crystal structure of R60E mutant of the histidine kinase (KinB) sensor domain from Pseudomonas aeruginosa PA01
To be Published
|
|
