3V6K
 
 | | Replication of N2,3-Ethenoguanine by DNA Polymerases | | 分子名称: | CALCIUM ION, DNA (5'-D(*TP*CP*AP*CP*(EFG)P*GP*AP*AP*TP*CP*CP*TP*TP*C)-3'), DNA (5'-D(P*GP*AP*AP*GP*GP*AP*TP*TP*CP*(2DT))-3'), ... | | 著者 | Zhao, L. | | 登録日 | 2011-12-20 | | 公開日 | 2012-04-25 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (3.6 Å) | | 主引用文献 | Replication of n(2) ,3-ethenoguanine by DNA polymerases.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
3VA7
 
 | |
3VIO
 
 | | Crystal structure of beta-glucosidase from termite Neotermes koshunensis in complex with a new glucopyranosidic product | | 分子名称: | 3-{4-[2-(beta-D-glucopyranosyloxy)ethyl]piperazin-1-yl}propane-1-sulfonic acid, Beta-glucosidase, CHLORIDE ION, ... | | 著者 | Jeng, W.Y, Liu, C.I, Wang, A.H.J. | | 登録日 | 2011-10-03 | | 公開日 | 2012-07-04 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.12 Å) | | 主引用文献 | High-resolution structures of Neotermes koshunensis beta-glucosidase mutants provide insights into the catalytic mechanism and the synthesis of glucoconjugates
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3VII
 
 | | Crystal structure of beta-glucosidase from termite Neotermes koshunensis in complex with Bis-Tris | | 分子名称: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-glucosidase, GLYCEROL, ... | | 著者 | Jeng, W.Y, Liu, C.I, Wang, A.H.J. | | 登録日 | 2011-10-03 | | 公開日 | 2012-07-04 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (0.97 Å) | | 主引用文献 | High-resolution structures of Neotermes koshunensis beta-glucosidase mutants provide insights into the catalytic mechanism and the synthesis of glucoconjugates
Acta Crystallogr.,Sect.D, 68, 2012
|
|
1EMS
 
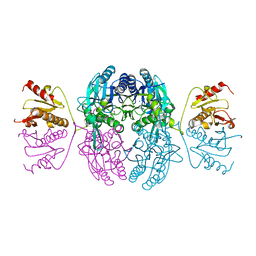 | | CRYSTAL STRUCTURE OF THE C. ELEGANS NITFHIT PROTEIN | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, ETHYL MERCURY ION, NIT-FRAGILE HISTIDINE TRIAD FUSION PROTEIN, ... | | 著者 | Pace, H.C, Hodawadekar, S.C, Draganescu, A, Huang, J, Bieganowski, P, Pekarsky, Y, Croce, C.M, Brenner, C. | | 登録日 | 2000-03-17 | | 公開日 | 2000-07-20 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal structure of the worm NitFhit Rosetta Stone protein reveals a Nit tetramer binding two Fhit dimers.
Curr.Biol., 10, 2000
|
|
1EUU
 
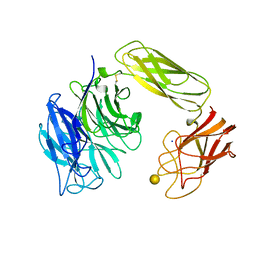 | | SIALIDASE OR NEURAMINIDASE, LARGE 68KD FORM | | 分子名称: | SIALIDASE, SODIUM ION, beta-D-galactopyranose | | 著者 | Gaskell, A, Crennell, S.J, Taylor, G.L. | | 登録日 | 1996-06-21 | | 公開日 | 1997-01-11 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | The three domains of a bacterial sialidase: a beta-propeller, an immunoglobulin module and a galactose-binding jelly-roll.
Structure, 3, 1995
|
|
3VIG
 
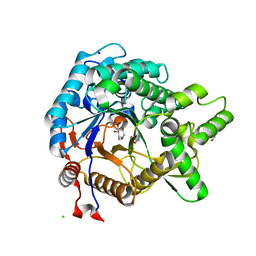 | | Crystal structure of beta-glucosidase from termite Neotermes koshunensis in complex with 1-deoxynojirimycin | | 分子名称: | 1-DEOXYNOJIRIMYCIN, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-glucosidase, ... | | 著者 | Jeng, W.Y, Liu, C.I, Wang, A.H.J. | | 登録日 | 2011-10-03 | | 公開日 | 2012-07-04 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (0.99 Å) | | 主引用文献 | High-resolution structures of Neotermes koshunensis beta-glucosidase mutants provide insights into the catalytic mechanism and the synthesis of glucoconjugates
Acta Crystallogr.,Sect.D, 68, 2012
|
|
2M0U
 
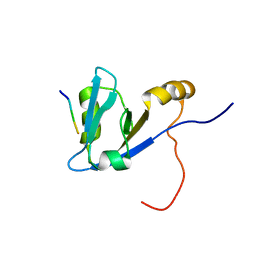 | | Complex structure of C-terminal CFTR peptide and extended PDZ1 domain from NHERF1 | | 分子名称: | C-terminal CFTR peptide, Na(+)/H(+) exchange regulatory cofactor NHE-RF1 | | 著者 | Bhattacharya, S, Ju, J.H, Cowburn, D, Bu, Z. | | 登録日 | 2012-11-06 | | 公開日 | 2013-04-24 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Ligand-Induced Dynamic Changes in Extended PDZ Domains from NHERF1.
J.Mol.Biol., 425, 2013
|
|
3VDG
 
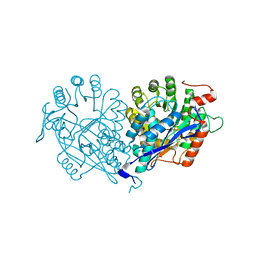 | | Crystal structure of enolase MSMEG_6132 (TARGET EFI-502282) from Mycobacterium smegmatis str. MC2 155 complexed with formate and acetate | | 分子名称: | ACETATE ION, CHLORIDE ION, FORMIC ACID, ... | | 著者 | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2012-01-05 | | 公開日 | 2012-01-25 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of enolase MSMEG_6132 FROM Mycobacterium smegmatis
To be Published
|
|
3VER
 
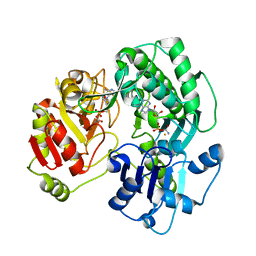 | | Crystal structure of the O-carbamoyltransferase TobZ in complex with carbamoyl adenylate intermediate | | 分子名称: | 5'-O-[(S)-(carbamoyloxy)(hydroxy)phosphoryl]adenosine, FE (II) ION, O-carbamoyltransferase TobZ, ... | | 著者 | Parthier, C, Stubbs, M.T, Goerlich, S, Jaenecke, F. | | 登録日 | 2012-01-09 | | 公開日 | 2012-01-25 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.34 Å) | | 主引用文献 | The O-Carbamoyltransferase TobZ Catalyzes an Ancient Enzymatic Reaction.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
2Z1N
 
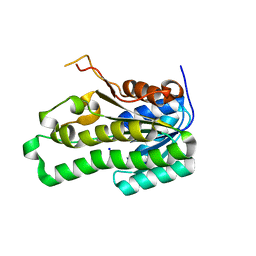 | | Crystal structure of APE0912 from Aeropyrum pernix K1 | | 分子名称: | SODIUM ION, dehydrogenase | | 著者 | Ichimura, T, Yamamura, A, Mimoto, F, Ohtsuka, J, Miyazono, K, Okai, M, Kamo, M, Lee, W.-C, Nagata, K, Tanokura, M. | | 登録日 | 2007-05-10 | | 公開日 | 2008-03-18 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | A unique catalytic triad revealed by the crystal structure of APE0912, a short-chain dehydrogenase/reductase family protein from Aeropyrum pernix K1
Proteins, 70, 2008
|
|
3VIH
 
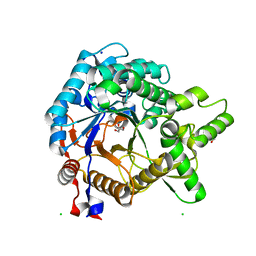 | | Crystal structure of beta-glucosidase from termite Neotermes koshunensis in complex with glycerol | | 分子名称: | Beta-glucosidase, CHLORIDE ION, GLYCEROL, ... | | 著者 | Jeng, W.Y, Liu, C.I, Wang, A.H.J. | | 登録日 | 2011-10-03 | | 公開日 | 2012-07-04 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.03 Å) | | 主引用文献 | High-resolution structures of Neotermes koshunensis beta-glucosidase mutants provide insights into the catalytic mechanism and the synthesis of glucoconjugates
Acta Crystallogr.,Sect.D, 68, 2012
|
|
2Z68
 
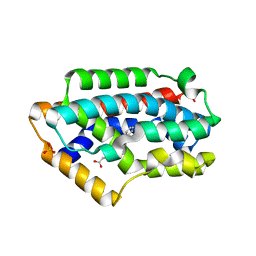 | | Crystal Structure Of An Artificial Metalloprotein: Cr[N-salicylidene-4-amino-3-hydroxyhydrocinnamic acid]/Wild Type Heme oxygenase | | 分子名称: | Heme oxygenase, SODIUM ION, SULFATE ION, ... | | 著者 | Yokoi, N, Unno, M, Ueno, T, Ikeda-Saito, M, Watanabe, Y. | | 登録日 | 2007-07-24 | | 公開日 | 2007-08-28 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.58 Å) | | 主引用文献 | Ligand design for improvement of thermal stabilityof metal complex/protein hybrids
To be Published
|
|
2MAT
 
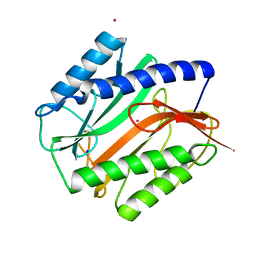 | | E.COLI METHIONINE AMINOPEPTIDASE AT 1.9 ANGSTROM RESOLUTION | | 分子名称: | COBALT (II) ION, PROTEIN (METHIONINE AMINOPEPTIDASE), SODIUM ION | | 著者 | Lowther, W.T, Orville, A.M, Madden, D.T, Lim, S, Rich, D.H, Matthews, B.W. | | 登録日 | 1999-03-29 | | 公開日 | 1999-06-18 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Escherichia coli methionine aminopeptidase: implications of crystallographic analyses of the native, mutant, and inhibited enzymes for the mechanism of catalysis.
Biochemistry, 38, 1999
|
|
2ZDV
 
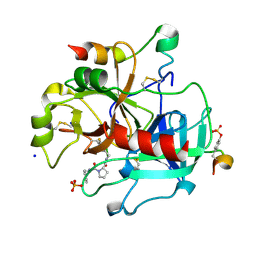 | | Exploring Thrombin S1 pocket | | 分子名称: | D-phenylalanyl-N-(3-fluorobenzyl)-L-prolinamide, Hirudin variant-1, PHOSPHATE ION, ... | | 著者 | Baum, B, Heine, A, Klebe, G. | | 登録日 | 2007-11-29 | | 公開日 | 2008-10-28 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.72 Å) | | 主引用文献 | Exploring Thrombin S1 pocket
To be Published
|
|
2ZHE
 
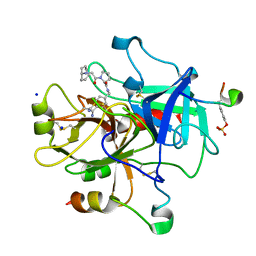 | | Exploring thrombin S3 pocket | | 分子名称: | Hirudin variant-2, N-cyclooctylglycyl-N-(4-carbamimidoylbenzyl)-L-prolinamide, SODIUM ION, ... | | 著者 | Brandt, T, Baum, B, Heine, A, Klebe, G. | | 登録日 | 2008-02-04 | | 公開日 | 2009-02-10 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Exploring thrombin S3 pocket
To be Published
|
|
3VQJ
 
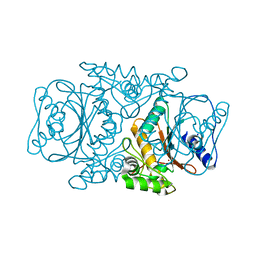 | | Crystal Structutre of Thiobacillus thioparus THI115 Carbonyl Sulfide Hydrolase | | 分子名称: | Carbonyl sulfide hydrolase, SODIUM ION, ZINC ION | | 著者 | Katayama, Y, Noguchi, K, Ogawa, T, Ohtaki, A, Odaka, M, Yohda, M. | | 登録日 | 2012-03-24 | | 公開日 | 2013-02-27 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Carbonyl Sulfide Hydrolase from Thiobacillus thioparus Strain THI115 Is One of the beta-Carbonic Anhydrase Family Enzymes
J.Am.Chem.Soc., 135, 2013
|
|
3VFA
 
 | | Crystal Structure of HIV-1 Protease Mutant V82A with novel P1'-Ligands GRL-02031 | | 分子名称: | (3aS,5R,6aR)-hexahydro-2H-cyclopenta[b]furan-5-yl [(1S,2R)-1-benzyl-2-hydroxy-3-([(4-methoxyphenyl)sulfonyl]{[(2R)-5-oxopyrrolidin-2-yl]methyl}amino)propyl]carbamate, CHLORIDE ION, SODIUM ION, ... | | 著者 | Yu, X.X, Wang, Y.F, Chang, Y.C.E, Weber, I.T. | | 登録日 | 2012-01-09 | | 公開日 | 2012-11-21 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.43 Å) | | 主引用文献 | Potent antiviral HIV-1 protease inhibitor GRL-02031 adapts to the structures of drug resistant mutants with its P1'-pyrrolidinone ring.
J.Med.Chem., 55, 2012
|
|
2ZGX
 
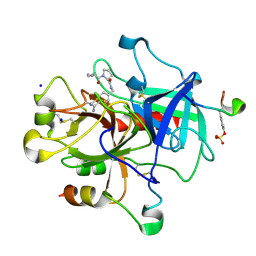 | | Thrombin Inhibition | | 分子名称: | 1-[(2R)-2-aminobutanoyl]-N-(4-carbamimidoylbenzyl)-L-prolinamide, Hirudin variant-1, SODIUM ION, ... | | 著者 | Baum, B, Heine, A, Klebe, G. | | 登録日 | 2008-01-28 | | 公開日 | 2008-12-16 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Think twice: understanding the high potency of bis(phenyl)methane inhibitors of thrombin
J.Mol.Biol., 391, 2009
|
|
3VI2
 
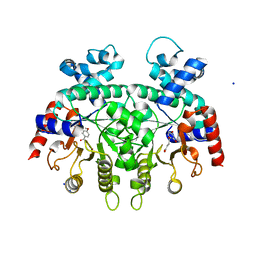 | | Crystal Structure Analysis of Plasmodium falciparum OMP Decarboxylase in complex with inhibitor HMOA | | 分子名称: | 4-(2-hydroxy-4-methoxyphenyl)-4-oxobutanoic acid, Orotidine 5'-phosphate decarboxylase, SODIUM ION | | 著者 | Takashima, Y, Mizohata, E, Krungkrai, S.R, Matsumura, H, Krungkrai, J, Horii, T, Inoue, T. | | 登録日 | 2011-09-16 | | 公開日 | 2012-08-01 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | The in silico screening and X-ray structure analysis of the inhibitor complex of Plasmodium falciparum orotidine 5'-monophosphate decarboxylase
J.Biochem., 152, 2012
|
|
3V7Z
 
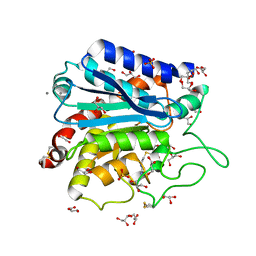 | | Carboxypeptidase T with GEMSA | | 分子名称: | (2-GUANIDINOETHYLMERCAPTO)SUCCINIC ACID, CALCIUM ION, Carboxypeptidase T, ... | | 著者 | Kuznetsov, S.A, Timofeev, V.I, Akparov, V.K, Kuranova, I.P. | | 登録日 | 2011-12-22 | | 公開日 | 2012-12-26 | | 最終更新日 | 2015-09-30 | | 実験手法 | X-RAY DIFFRACTION (1.61 Å) | | 主引用文献 | Structural insights into the broad substrate specificity of carboxypeptidase T from Thermoactinomyces vulgaris.
Febs J., 282, 2015
|
|
3VRK
 
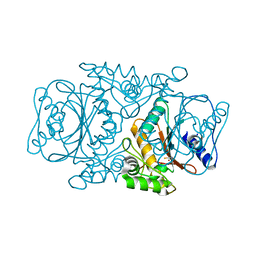 | | Crystal Structutre of Thiobacillus thioparus THI115 Carbonyl Sulfide Hydrolase / Thiocyanate complex | | 分子名称: | Carbonyl sulfide hydrolase, SODIUM ION, THIOCYANATE ION, ... | | 著者 | Katayama, Y, Noguchi, K, Ogawa, T, Ohtaki, A, Odaka, M, Yohda, M. | | 登録日 | 2012-04-11 | | 公開日 | 2013-02-27 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.33 Å) | | 主引用文献 | Carbonyl Sulfide Hydrolase from Thiobacillus thioparus Strain THI115 Is One of the beta-Carbonic Anhydrase Family Enzymes
J.Am.Chem.Soc., 135, 2013
|
|
2M0V
 
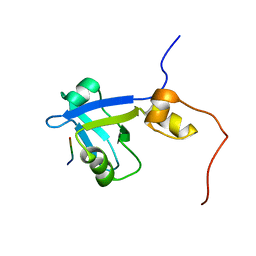 | | Complex structure of C-terminal CFTR peptide and extended PDZ2 domain from NHERF1 | | 分子名称: | C-terminal CFTR peptide, Na(+)/H(+) exchange regulatory cofactor NHE-RF1 | | 著者 | Bhattacharya, S, Ju, J.H, Cowburn, D, Bu, Z. | | 登録日 | 2012-11-06 | | 公開日 | 2013-04-24 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Ligand-Induced Dynamic Changes in Extended PDZ Domains from NHERF1.
J.Mol.Biol., 425, 2013
|
|
3VE2
 
 | |
3VFB
 
 | | Crystal Structure of HIV-1 Protease Mutant N88D with novel P1'-Ligands GRL-02031 | | 分子名称: | (3aS,5R,6aR)-hexahydro-2H-cyclopenta[b]furan-5-yl [(1S,2R)-1-benzyl-2-hydroxy-3-([(4-methoxyphenyl)sulfonyl]{[(2R)-5-oxopyrrolidin-2-yl]methyl}amino)propyl]carbamate, CHLORIDE ION, GLYCEROL, ... | | 著者 | Yu, X.X, Wang, Y.F, Chang, Y.C.E, Weber, I.T. | | 登録日 | 2012-01-09 | | 公開日 | 2012-11-21 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Potent antiviral HIV-1 protease inhibitor GRL-02031 adapts to the structures of drug resistant mutants with its P1'-pyrrolidinone ring.
J.Med.Chem., 55, 2012
|
|
