3UYC
 
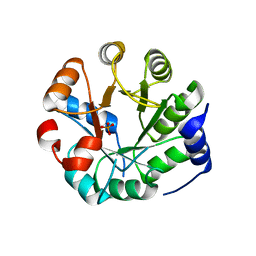 | | Designed protein KE59 R8_2/7A | | 分子名称: | Kemp eliminase KE59 R8_2/7A, PHOSPHATE ION | | 著者 | Khersonsky, O, Kiss, G, Roethlisberger, D, Dym, O, Albeck, S, Houk, K.N, Baker, D, Tawfik, D.S, Israel Structural Proteomics Center (ISPC) | | 登録日 | 2011-12-06 | | 公開日 | 2012-06-06 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Bridging the gaps in design methodologies by evolutionary optimization of the stability and proficiency of designed Kemp eliminase KE59.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3UXD
 
 | | Designed protein KE59 R1 7/10H with dichlorobenzotriazole (DBT) | | 分子名称: | 5,7-dichloro-1H-benzotriazole, Kemp eliminase KE59 R1 7/10H, PHOSPHATE ION | | 著者 | Khersonsky, O, Kiss, G, Roethlisberger, D, Dym, O, Albeck, S, Houk, K.N, Baker, D, Tawfik, D.S, Israel Structural Proteomics Center (ISPC) | | 登録日 | 2011-12-05 | | 公開日 | 2012-06-06 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Bridging the gaps in design methodologies by evolutionary optimization of the stability and proficiency of designed Kemp eliminase KE59.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3UY7
 
 | | Designed protein KE59 R1 7/10H with G130S mutation | | 分子名称: | Kemp eliminase KE59 R1 7/10H, SODIUM ION, SULFATE ION | | 著者 | Khersonsky, O, Kiss, G, Roethlisberger, D, Dym, O, Albeck, S, Houk, K.N, Baker, D, Tawfik, D.S, Israel Structural Proteomics Center (ISPC) | | 登録日 | 2011-12-06 | | 公開日 | 2012-06-27 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Bridging the gaps in design methodologies by evolutionary optimization of the stability and proficiency of designed Kemp eliminase KE59.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3UXA
 
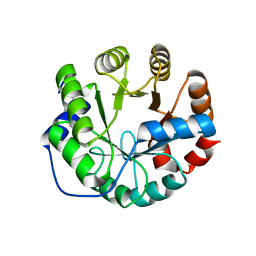 | | Designed protein KE59 R1 7/10H | | 分子名称: | Kemp eliminase KE59 R1 7/10H, PHOSPHATE ION | | 著者 | Khersonsky, O, Kiss, G, Roethlisberger, D, Dym, O, Albeck, S, Houk, K.N, Baker, D, Tawfik, D.S, Israel Structural Proteomics Center (ISPC) | | 登録日 | 2011-12-05 | | 公開日 | 2012-06-06 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Bridging the gaps in design methodologies by evolutionary optimization of the stability and proficiency of designed Kemp eliminase KE59.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3UZ5
 
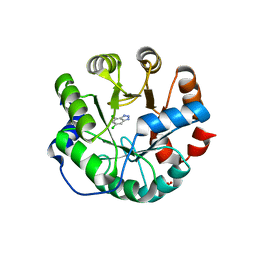 | | Designed protein KE59 R13 3/11H | | 分子名称: | 5,7-dichloro-1H-benzotriazole, Kemp eliminase KE59 R13 3/11H, PHOSPHATE ION, ... | | 著者 | Khersonsky, O, Kiss, G, Roethlisberger, D, Dym, O, Albeck, S, Houk, K.N, Baker, D, Tawfik, D.S, Israel Structural Proteomics Center (ISPC) | | 登録日 | 2011-12-07 | | 公開日 | 2012-06-06 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Bridging the gaps in design methodologies by evolutionary optimization of the stability and proficiency of designed Kemp eliminase KE59.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3UZJ
 
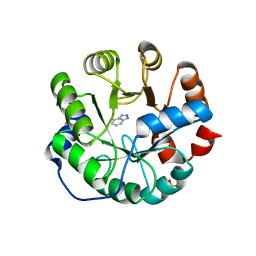 | | Designed protein KE59 R13 3/11H with benzotriazole | | 分子名称: | 1H-benzotriazole, Kemp eliminase KE59 R13 3/11H, PHOSPHATE ION | | 著者 | Khersonsky, O, Kiss, G, Roethlisberger, D, Dym, O, Albeck, S, Houk, K.N, Baker, D, Tawfik, D.S, Israel Structural Proteomics Center (ISPC) | | 登録日 | 2011-12-07 | | 公開日 | 2012-06-06 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.69 Å) | | 主引用文献 | Bridging the gaps in design methodologies by evolutionary optimization of the stability and proficiency of designed Kemp eliminase KE59.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3UY8
 
 | | Designed protein KE59 R5_11/5F | | 分子名称: | Kemp eliminase KE59 R5_11/5F, SULFATE ION | | 著者 | Khersonsky, O, Kiss, G, Roethlisberger, D, Dym, O, Albeck, S, Houk, K.N, Baker, D, Tawfik, D.S, Israel Structural Proteomics Center (ISPC) | | 登録日 | 2011-12-06 | | 公開日 | 2012-06-06 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.41 Å) | | 主引用文献 | Bridging the gaps in design methodologies by evolutionary optimization of the stability and proficiency of designed Kemp eliminase KE59.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
7JG3
 
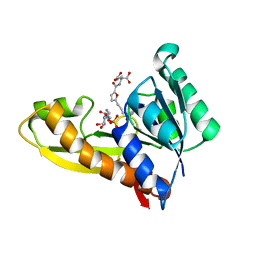 | | Human GAR transformylase in complex with GAR substrate and AGF103 inhibitor | | 分子名称: | GLYCINAMIDE RIBONUCLEOTIDE, N-{5-[4-(2-amino-4-oxo-3,4-dihydrothieno[2,3-d]pyrimidin-6-yl)butyl]furan-2-carbonyl}-L-glutamic acid, SODIUM ION, ... | | 著者 | Wong-Roushar, J, Dann III, C.E. | | 登録日 | 2020-07-18 | | 公開日 | 2021-03-31 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.091 Å) | | 主引用文献 | Discovery of 6-substituted thieno[2,3-d]pyrimidine analogs as dual inhibitors of glycinamide ribonucleotide formyltransferase and 5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase in de novo purine nucleotide biosynthesis in folate receptor expressing human tumors
Bioorg.Med.Chem., 37, 2021
|
|
7JG4
 
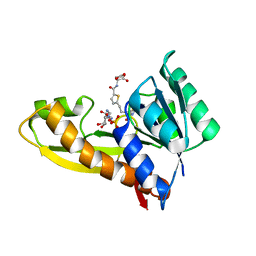 | | Human GAR transformylase in complex with GAR substrate and AGF131 inhibitor | | 分子名称: | GLYCINAMIDE RIBONUCLEOTIDE, N-(5-{3-[(1S,7R,8R,9S)-4-amino-2-oxo-7lambda~4~-thia-3,5-diazatetracyclo[4.3.0.0~1,7~.0~7,9~]nona-3,5-dien-8-yl]propyl}thiophene-2-carbonyl)-L-glutamic acid, SODIUM ION, ... | | 著者 | Wong-Roushar, J, Dann III, C.E. | | 登録日 | 2020-07-18 | | 公開日 | 2021-03-31 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.455 Å) | | 主引用文献 | Discovery of 6-substituted thieno[2,3-d]pyrimidine analogs as dual inhibitors of glycinamide ribonucleotide formyltransferase and 5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase in de novo purine nucleotide biosynthesis in folate receptor expressing human tumors
Bioorg.Med.Chem., 37, 2021
|
|
7JG0
 
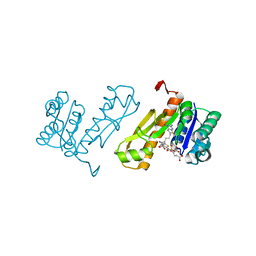 | | Human GAR transformylase in complex with GAR substrate and AGF102 inhibitor | | 分子名称: | GLYCINAMIDE RIBONUCLEOTIDE, N-{5-[4-(2-amino-4-oxo-3,4-dihydrothieno[2,3-d]pyrimidin-6-yl)butyl]thiophene-2-carbonyl}-L-glutamic acid, Trifunctional purine biosynthetic protein adenosine-3 | | 著者 | Wong-Roushar, J, Dann III, C.E. | | 登録日 | 2020-07-18 | | 公開日 | 2021-03-31 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.984 Å) | | 主引用文献 | Discovery of 6-substituted thieno[2,3-d]pyrimidine analogs as dual inhibitors of glycinamide ribonucleotide formyltransferase and 5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase in de novo purine nucleotide biosynthesis in folate receptor expressing human tumors
Bioorg.Med.Chem., 37, 2021
|
|
2LVB
 
 | | Solution NMR Structure DE NOVO DESIGNED PFK fold PROTEIN, Northeast Structural Genomics Consortium (NESG) Target OR250 | | 分子名称: | DE NOVO DESIGNED PFK fold PROTEIN | | 著者 | Liu, G, Koga, N, Koga, R, Xiao, R, Hamilton, K, Kohan, E, Acton, T.B, Kornhaber, G, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2012-06-30 | | 公開日 | 2012-08-08 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Principles for designing ideal protein structures.
Nature, 491, 2012
|
|
2LV8
 
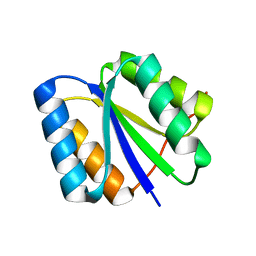 | | Solution NMR Structure de novo designed rossmann 2x2 fold protein, Northeast Structural Genomics Consortium (NESG) Target OR16 | | 分子名称: | De novo designed rossmann 2x2 fold protein | | 著者 | Liu, G, Koga, R, Koga, N, Xiao, R, Pederson, K, Hamilton, K, Ciccosanti, C, Acton, T.B, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2012-06-29 | | 公開日 | 2012-08-08 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Principles for designing ideal protein structures.
Nature, 491, 2012
|
|
8D9O
 
 | | De Novo Photosynthetic Reaction Center Protein in Apo-State | | 分子名称: | CADMIUM ION, Reaction Center Maquette | | 著者 | Ennist, N.M, Stayrook, S.E, Dutton, P.L, Moser, C.C. | | 登録日 | 2022-06-10 | | 公開日 | 2022-09-28 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | Rational design of photosynthetic reaction center protein maquettes.
Front Mol Biosci, 9, 2022
|
|
8D9P
 
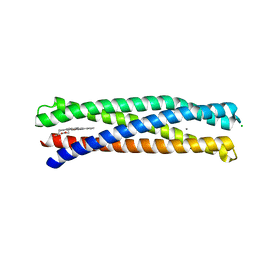 | | De Novo Photosynthetic Reaction Center Protein Equipped with Heme B and Mn(II) cations | | 分子名称: | CHLORIDE ION, MANGANESE (II) ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | 著者 | Ennist, N.M, Stayrook, S.E, Dutton, P.L, Moser, C.C. | | 登録日 | 2022-06-10 | | 公開日 | 2022-09-28 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Rational design of photosynthetic reaction center protein maquettes.
Front Mol Biosci, 9, 2022
|
|
3S0R
 
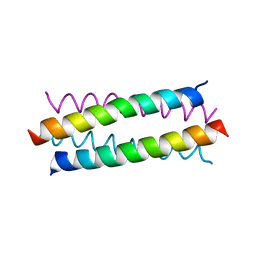 | |
4E88
 
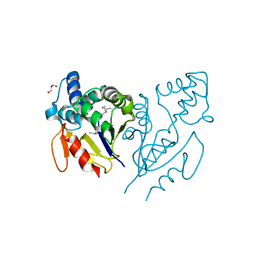 | | CRYSTAL STRUCTURE OF DE NOVO DESIGNED CYSTEINE ESTERASE ECH13, Northeast Structural Genomics Consortium Target OR51 | | 分子名称: | DE NOVO DESIGNED CYSTEINE ESTERASE ECH13, GLYCEROL | | 著者 | Kuzin, A, Su, M, Seetharaman, J, Xiao, X, Sahdev, S, Ciccosanti, C, Richter, F, Everett, J.K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2012-03-19 | | 公開日 | 2012-04-11 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Northeast Structural Genomics Consortium Target OR51
To be Published
|
|
1NVO
 
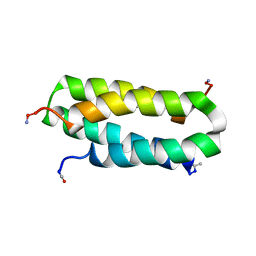 | | Solution structure of a four-helix bundle model, apo-DF1 | | 分子名称: | Homodimeric Alpha2 Four-Helix Bundle | | 著者 | Maglio, O, Nastri, F, Pavone, V, Lombardi, A, DeGrado, W.F. | | 登録日 | 2003-02-04 | | 公開日 | 2003-03-25 | | 最終更新日 | 2022-02-23 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Preorganization of molecular binding sites in designed diiron proteins
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
3U1V
 
 | | X-ray Structure of De Novo design cysteine esterase FR29, Northeast Structural Genomics Consortium Target OR52 | | 分子名称: | De Novo design cysteine esterase FR29 | | 著者 | Kuzin, A, Su, M, Vorobiev, S.M, Seetharaman, J, Patel, D, Xiao, R, Ciccosanti, C, Richter, F, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2011-09-30 | | 公開日 | 2011-12-07 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.797 Å) | | 主引用文献 | Computational design of catalytic dyads and oxyanion holes for ester hydrolysis.
J.Am.Chem.Soc., 134, 2012
|
|
3UAK
 
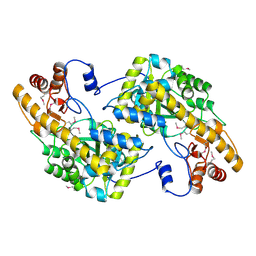 | | Crystal Structure of De Novo designed cysteine esterase ECH14, Northeast Structural Genomics Consortium Target OR54 | | 分子名称: | De Novo designed cysteine esterase ECH14 | | 著者 | Kuzin, A, Su, M, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Richter, F, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Baker, D, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2011-10-21 | | 公開日 | 2011-12-07 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (3.232 Å) | | 主引用文献 | Computational design of catalytic dyads and oxyanion holes for ester hydrolysis.
J.Am.Chem.Soc., 134, 2012
|
|
3U1O
 
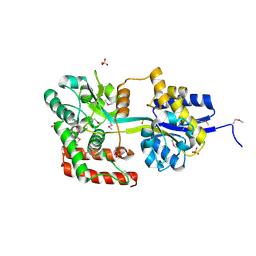 | | THREE DIMENSIONAL STRUCTURE OF DE NOVO DESIGNED CYSTEINE ESTERASE ECH19, Northeast Structural Genomics Consortium Target OR49 | | 分子名称: | De Novo design cysteine esterase ECH19, SODIUM ION, SULFATE ION | | 著者 | Kuzin, A, Su, M, Lew, S, Forouhar, F, Seetharaman, J, Daya, P, Xiao, R, Ciccosanti, C, Richter, F, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2011-09-30 | | 公開日 | 2011-10-26 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.494 Å) | | 主引用文献 | Computational design of catalytic dyads and oxyanion holes for ester hydrolysis.
J.Am.Chem.Soc., 134, 2012
|
|
2LQ1
 
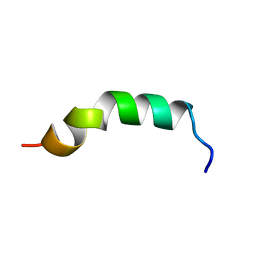 | |
2LQ0
 
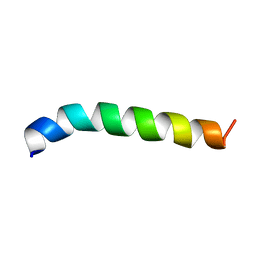 | |
2LQ2
 
 | | Solution structure of de novo designed peptide 4m | | 分子名称: | de novo designed antifreeze peptide 4m | | 著者 | Bhunia, A. | | 登録日 | 2012-02-22 | | 公開日 | 2012-10-24 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structures, dynamics, and ice growth inhibitory activity of Peptide fragments derived from an antarctic yeast protein
Plos One, 7, 2012
|
|
2KDM
 
 | | NMR structures of GA95 and GB95, two designed proteins with 95% sequence identity but different folds and functions | | 分子名称: | DESIGNED PROTEIN | | 著者 | He, Y, Alexander, P, Chen, Y, Bryan, P, Orban, J. | | 登録日 | 2009-01-12 | | 公開日 | 2009-12-29 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | From the Cover: A minimal sequence code for switching protein structure and function.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2KDL
 
 | | NMR structures of GA95 and GB95, two designed proteins with 95% sequence identity but different folds and functions | | 分子名称: | designed protein | | 著者 | He, Y, Alexander, P, Chen, Y, Bryan, P, Orban, J. | | 登録日 | 2009-01-12 | | 公開日 | 2009-12-29 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | A minimal sequence code for switching protein structure and function.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
