7LCE
 
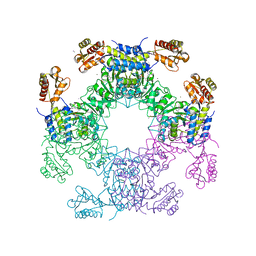 | | Structure of D-Glucosaminate-6-phosphate Ammonia-lyase | | 分子名称: | CALCIUM ION, D-glucosaminate-6-phosphate ammonia lyase, DIMETHYL SULFOXIDE | | 著者 | Phillips, R.S. | | 登録日 | 2021-01-10 | | 公開日 | 2021-06-09 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.58 Å) | | 主引用文献 | Structure and Mechanism of d-Glucosaminate-6-phosphate Ammonia-lyase: A Novel Octameric Assembly for a Pyridoxal 5'-Phosphate-Dependent Enzyme, and Unprecedented Stereochemical Inversion in the Elimination Reaction of a d-Amino Acid.
Biochemistry, 60, 2021
|
|
7S76
 
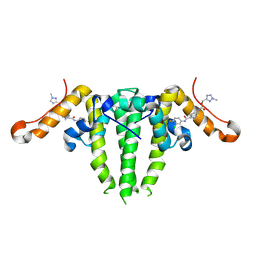 | | HBV CAPSID Y132A WITH COMPOUND 10b AT 2.5A RESOLUTION | | 分子名称: | (1-methyl-1H-1,2,4-triazol-3-yl)methyl {(4S)-1-[(3-chloro-4-fluorophenyl)carbamoyl]-2-methyl-2,4,5,6-tetrahydrocyclopenta[c]pyrrol-4-yl}carbamate, Capsid protein | | 著者 | Olland, A.M, Suto, R.K. | | 登録日 | 2021-09-15 | | 公開日 | 2022-05-04 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | The identification of highly efficacious functionalised tetrahydrocyclopenta[ c ]pyrroles as inhibitors of HBV viral replication through modulation of HBV capsid assembly.
Rsc Med Chem, 13, 2022
|
|
1L82
 
 | |
6AT1
 
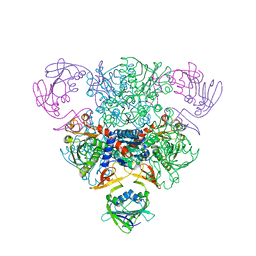 | | STRUCTURAL CONSEQUENCES OF EFFECTOR BINDING TO THE T STATE OF ASPARTATE CARBAMOYLTRANSFERASE. CRYSTAL STRUCTURES OF THE UNLIGATED AND ATP-, AND CTP-COMPLEXED ENZYMES AT 2.6-ANGSTROMS RESOLUTION | | 分子名称: | ASPARTATE CARBAMOYLTRANSFERASE (T STATE), CATALYTIC CHAIN, ASPARTATE CARBAMOYLTRANSFERASE REGULATORY CHAIN, ... | | 著者 | Stevens, R.C, Gouaux, J.E, Lipscomb, W.N. | | 登録日 | 1990-04-26 | | 公開日 | 1990-10-15 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural consequences of effector binding to the T state of aspartate carbamoyltransferase: crystal structures of the unligated and ATP- and CTP-complexed enzymes at 2.6-A resolution.
Biochemistry, 29, 1990
|
|
7NJ0
 
 | | CryoEM structure of the human Separase-Cdk1-cyclin B1-Cks1 complex | | 分子名称: | Cyclin-dependent kinase 1, Cyclin-dependent kinases regulatory subunit 1, G2/mitotic-specific cyclin-B1,G2/mitotic-specific cyclin-B1, ... | | 著者 | Yu, J, Raia, P, Ghent, C.M, Raisch, T, Sadian, Y, Barford, D, Raunser, S, Morgan, D.O, Boland, A. | | 登録日 | 2021-02-14 | | 公開日 | 2021-08-04 | | 最終更新日 | 2021-08-18 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structural basis of human separase regulation by securin and CDK1-cyclin B1.
Nature, 596, 2021
|
|
6C8S
 
 | | Loganic acid methyltransferase with SAH | | 分子名称: | Loganic acid O-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | 著者 | Petronikolou, N, Nair, S.K. | | 登録日 | 2018-01-25 | | 公開日 | 2018-02-21 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Loganic Acid Methyltransferase: Insights into the Specificity of Methylation on an Iridoid Glycoside.
Chembiochem, 19, 2018
|
|
8A60
 
 | |
7ZQK
 
 | | Crystal structure of photosynthetic glyceraldehyde-3-phosphate dehydrogenase from Chlamydomonas reinhardtii (CrGAPA) complexed with NAD+ | | 分子名称: | Glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | 著者 | Fermani, S, Zaffagnini, M, Lemaire, S.D, Falini, G, Fanti, S, Rossi, J. | | 登録日 | 2022-04-30 | | 公開日 | 2022-07-20 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural snapshots of nitrosoglutathione binding and reactivity underlying S-nitrosylation of photosynthetic GAPDH.
Redox Biol, 54, 2022
|
|
7ZQ4
 
 | | Crystal structure of photosynthetic glyceraldehyde-3-phosphate dehydrogenase from Chlamydomonas reinhardtii (CrGAPA) complexed with NADP+ and the oxidated catalytic cysteine | | 分子名称: | 1,2-ETHANEDIOL, GLYCEROL, Glyceraldehyde-3-phosphate dehydrogenase A, ... | | 著者 | Fermani, S, Zaffagnini, M, Lemaire, S.D, Falini, G, Fanti, S, Rossi, J. | | 登録日 | 2022-04-29 | | 公開日 | 2022-07-20 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural snapshots of nitrosoglutathione binding and reactivity underlying S-nitrosylation of photosynthetic GAPDH.
Redox Biol, 54, 2022
|
|
7ZQ3
 
 | | Crystal structure of photosynthetic glyceraldehyde-3-phosphate dehydrogenase from Chlamydomonas reinhardtii (CrGAPA) complexed with NADP+ | | 分子名称: | Glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | 著者 | Fermani, S, Zaffagnini, M, Lemaire, S.D, Falini, G, Fanti, S, Rossi, J. | | 登録日 | 2022-04-29 | | 公開日 | 2022-07-20 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structural snapshots of nitrosoglutathione binding and reactivity underlying S-nitrosylation of photosynthetic GAPDH.
Redox Biol, 54, 2022
|
|
7NWH
 
 | | Mammalian pre-termination 80S ribosome with eRF1 and eRF3 bound by Blasticidin S. | | 分子名称: | 18S Ribosomal RNA, 28S Ribosomal RNA, 40S ribosomal protein S10, ... | | 著者 | Powers, K.T, Yadav, S.K.N, Bufton, J.C, Schaffitzel, C. | | 登録日 | 2021-03-16 | | 公開日 | 2021-07-07 | | 最終更新日 | 2023-01-11 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Blasticidin S inhibits mammalian translation and enhances production of protein encoded by nonsense mRNA.
Nucleic Acids Res., 49, 2021
|
|
9B5W
 
 | |
9B56
 
 | |
9B5J
 
 | |
9B5P
 
 | |
9B5E
 
 | |
8Q5B
 
 | |
9B5R
 
 | |
9CVG
 
 | |
9C0J
 
 | |
9B58
 
 | |
9BJK
 
 | | Inactive mu opioid receptor bound to Nb6, naloxone and NAM | | 分子名称: | Mu-type opioid receptor, Naloxone, Nalpha-[({(1M)-1-[5-(benzyloxy)pyridin-3-yl]naphthalen-2-yl}sulfanyl)acetyl]-3-methoxy-N,4-dimethyl-L-phenylalaninamide, ... | | 著者 | O'Brien, E.S, Wang, H, Kaavya Krishna, K, Zhang, C, Kobilka, B.K. | | 登録日 | 2024-04-25 | | 公開日 | 2024-07-17 | | 最終更新日 | 2024-08-14 | | 実験手法 | ELECTRON MICROSCOPY (3.26 Å) | | 主引用文献 | A mu-opioid receptor modulator that works cooperatively with naloxone.
Nature, 631, 2024
|
|
9B5I
 
 | |
9MHT
 
 | | CYTOSINE-SPECIFIC METHYLTRANSFERASE HHAI/DNA COMPLEX | | 分子名称: | 5'-D(P*CP*CP*AP*TP*GP*CP*GP*CP*TP*GP*AP*C)-3', 5'-D(P*GP*TP*CP*AP*GP*(3DR)P*GP*CP*AP*TP*GP*G)-3', CYTOSINE-SPECIFIC METHYLTRANSFERASE HHAI, ... | | 著者 | O'Gara, M, Horton, J.R, Roberts, R.J, Cheng, X. | | 登録日 | 1998-08-07 | | 公開日 | 1998-12-01 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.39 Å) | | 主引用文献 | Structures of HhaI methyltransferase complexed with substrates containing mismatches at the target base.
Nat.Struct.Biol., 5, 1998
|
|
9B5D
 
 | |
