1L80
 
 | |
1L00
 
 | | PERTURBATION OF TRP 138 IN T4 LYSOZYME BY MUTATIONS AT GLN 105 USED TO CORRELATE CHANGES IN STRUCTURE, STABILITY, SOLVATION, AND SPECTROSCOPIC PROPERTIES | | 分子名称: | BETA-MERCAPTOETHANOL, T4 LYSOZYME | | 著者 | Pjura, P, Mcintosh, L.P, Wozniak, J.A, Matthews, B.W. | | 登録日 | 1992-07-13 | | 公開日 | 1993-10-31 | | 最終更新日 | 2024-06-05 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Perturbation of Trp 138 in T4 lysozyme by mutations at Gln 105 used to correlate changes in structure, stability, solvation, and spectroscopic properties.
Proteins, 15, 1993
|
|
1L16
 
 | |
1L24
 
 | |
1L34
 
 | |
1L18
 
 | |
1L81
 
 | |
7QI3
 
 | | Structure of Fusarium verticillioides NAT1 (FDB2) N-malonyltransferase | | 分子名称: | 1,2-ETHANEDIOL, Arylamine N-acetyltransferase, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Lowe, E.D, Kotomina, E, Karagianni, E, Boukouvala, S. | | 登録日 | 2021-12-14 | | 公開日 | 2022-11-23 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Fusarium verticillioides NAT1 (FDB2) N-malonyltransferase is structurally, functionally and phylogenetically distinct from its N-acetyltransferase (NAT) homologues.
Febs J., 290, 2023
|
|
6ZB9
 
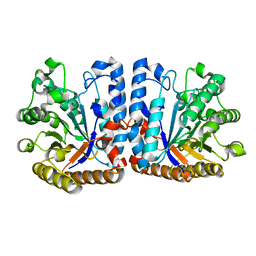 | | Exo-beta-1,3-glucanase from moose rumen microbiome, wild type | | 分子名称: | Exo-beta-1,3-glucanase | | 著者 | Kalyani, D.C, Reichenbach, T, Aspeborg, H, Divne, C. | | 登録日 | 2020-06-08 | | 公開日 | 2021-01-13 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | A homodimeric bacterial exo-beta-1,3-glucanase derived from moose rumen microbiome shows a structural framework similar to yeast exo-beta-1,3-glucanases.
Enzyme.Microb.Technol., 143, 2021
|
|
6ZB8
 
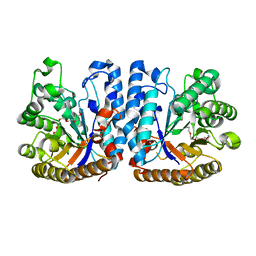 | | Exo-beta-1,3-glucanase from moose rumen microbiome, active site mutant E167Q/E295Q | | 分子名称: | Exo-beta-1,3-glucanase variant E167Q/E295Q, POLYETHYLENE GLYCOL (N=34) | | 著者 | Kalyani, D.C, Reichenbach, T, Aspeborg, H, Divne, C. | | 登録日 | 2020-06-08 | | 公開日 | 2021-01-13 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | A homodimeric bacterial exo-beta-1,3-glucanase derived from moose rumen microbiome shows a structural framework similar to yeast exo-beta-1,3-glucanases.
Enzyme.Microb.Technol., 143, 2021
|
|
7XRN
 
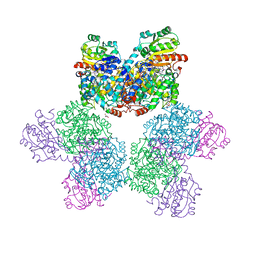 | | Ethanolamine ammonia-lyase complexed with AdoMeCbl in the presence of substrate | | 分子名称: | (2~{R},3~{R},4~{S},5~{R})-2-(6-aminopurin-9-yl)-5-ethyl-oxolane-3,4-diol, AMMONIUM ION, COBALAMIN, ... | | 著者 | Shibata, N, Toraya, T. | | 登録日 | 2022-05-10 | | 公開日 | 2023-03-29 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.07 Å) | | 主引用文献 | Structural Insights into the Very Low Activity of the Homocoenzyme B 12 Adenosylmethylcobalamin in Coenzyme B 12 -Dependent Diol Dehydratase and Ethanolamine Ammonia-Lyase.
Chemistry, 28, 2022
|
|
7XRK
 
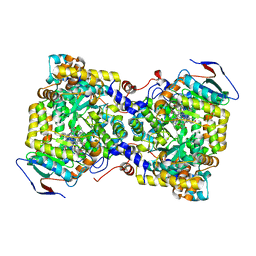 | | Diol dehydratase complexed with AdoMeCbl | | 分子名称: | (2~{R},3~{R},4~{S},5~{R})-2-(6-aminopurin-9-yl)-5-ethyl-oxolane-3,4-diol, CALCIUM ION, COBALAMIN, ... | | 著者 | Shibata, N, Toraya, T. | | 登録日 | 2022-05-10 | | 公開日 | 2023-03-29 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural Insights into the Very Low Activity of the Homocoenzyme B 12 Adenosylmethylcobalamin in Coenzyme B 12 -Dependent Diol Dehydratase and Ethanolamine Ammonia-Lyase.
Chemistry, 28, 2022
|
|
7XRM
 
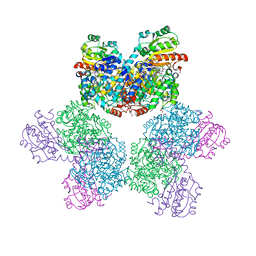 | | Ethanolamine ammonia-lyase complexed with AdoMeCbl | | 分子名称: | (2~{R},3~{R},4~{S},5~{R})-2-(6-aminopurin-9-yl)-5-ethyl-oxolane-3,4-diol, AMMONIUM ION, COBALAMIN, ... | | 著者 | Shibata, N, Toraya, T. | | 登録日 | 2022-05-10 | | 公開日 | 2023-03-29 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.13 Å) | | 主引用文献 | Structural Insights into the Very Low Activity of the Homocoenzyme B 12 Adenosylmethylcobalamin in Coenzyme B 12 -Dependent Diol Dehydratase and Ethanolamine Ammonia-Lyase.
Chemistry, 28, 2022
|
|
7XRL
 
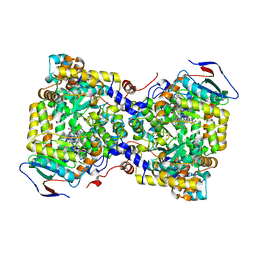 | | Diol dehydratase complexed with AdoMeCbl and 1,2-propanediol | | 分子名称: | (2~{R},3~{R},4~{S},5~{R})-2-(6-aminopurin-9-yl)-5-ethyl-oxolane-3,4-diol, AMMONIUM ION, CALCIUM ION, ... | | 著者 | Shibata, N, Toraya, T. | | 登録日 | 2022-05-10 | | 公開日 | 2023-03-29 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structural Insights into the Very Low Activity of the Homocoenzyme B 12 Adenosylmethylcobalamin in Coenzyme B 12 -Dependent Diol Dehydratase and Ethanolamine Ammonia-Lyase.
Chemistry, 28, 2022
|
|
6Z1H
 
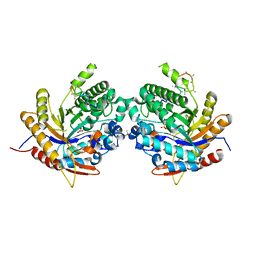 | | Ancestral glycosidase (family 1) | | 分子名称: | ANCESTRAL RECONSTRUCTED GLYCOSIDASE, GLYCEROL, ISOPROPYL ALCOHOL, ... | | 著者 | Gavira, J.A, Risso, V.A, Sanchez-Ruiz, J.M, Gamiz-Arco, G, Gutierrez-Rus, L, Ibarra-Molero, B, Hoshino, Y, Petrovic, D, Romero-Rivera, A, Seelig, B, Kamerlin, S.C.L, Gaucher, E.A. | | 登録日 | 2020-05-13 | | 公開日 | 2020-07-22 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Heme-binding enables allosteric modulation in an ancient TIM-barrel glycosidase.
Nat Commun, 12, 2021
|
|
7R7S
 
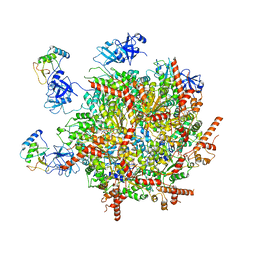 | | p47-bound p97-R155H mutant with ATPgammaS | | 分子名称: | NSFL1 cofactor p47, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Transitional endoplasmic reticulum ATPase | | 著者 | Nandi, P, Li, S, Coulmbres, R.C.A, Wang, F, Williams, D.R, Malyutin, A.G, Poh, Y.-P, Chou, T.-F, Chiu, P.-L. | | 登録日 | 2021-06-25 | | 公開日 | 2021-08-04 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (4.23 Å) | | 主引用文献 | Structural and Functional Analysis of Disease-Linked p97 ATPase Mutant Complexes.
Int J Mol Sci, 22, 2021
|
|
6Z1I
 
 | |
6Z1O
 
 | |
7RLH
 
 | | Cryo-EM structure of human p97-D592N mutant bound to ATPgS. | | 分子名称: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Transitional endoplasmic reticulum ATPase | | 著者 | Caffrey, B, Zhu, X, Berezuk, A, Tuttle, K, Chittori, S, Subramaniam, S. | | 登録日 | 2021-07-23 | | 公開日 | 2021-09-22 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | AAA+ ATPase p97/VCP mutants and inhibitor binding disrupt inter-domain coupling and subsequent allosteric activation.
J.Biol.Chem., 297, 2021
|
|
7P27
 
 | | NMR solution structure of Chikungunya virus macro domain | | 分子名称: | Polyprotein P1234 | | 著者 | Lykouras, M.V, Tsika, A.C, Papageorgiou, N, Canard, B, Coutard, B, Bentrop, D, Spyroulias, G.A. | | 登録日 | 2021-07-04 | | 公開日 | 2022-07-13 | | 最終更新日 | 2024-06-19 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Binding Adaptation of GS-441524 Diversifies Macro Domains and Downregulates SARS-CoV-2 de-MARylation Capacity.
J.Mol.Biol., 434, 2022
|
|
7RLF
 
 | | Cryo-EM structure of human p97-E470D mutant bound to ATPgS. | | 分子名称: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Transitional endoplasmic reticulum ATPase | | 著者 | Caffrey, B, Zhu, X, Berezuk, A, Tuttle, K, Chittori, S, Subramaniam, S. | | 登録日 | 2021-07-23 | | 公開日 | 2021-09-22 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | AAA+ ATPase p97/VCP mutants and inhibitor binding disrupt inter-domain coupling and subsequent allosteric activation.
J.Biol.Chem., 297, 2021
|
|
1L17
 
 | |
1L79
 
 | |
6YXH
 
 | | Cryogenic human alkaline ceramidase 3 (ACER3) at 2.6 A resolution determined by Serial Crystallography (SSX) using CrystalDirect | | 分子名称: | Alkaline ceramidase 3, CALCIUM ION, MAGNESIUM ION, ... | | 著者 | Healey, R.D, Basu, S, Humm, A.S, Leyrat, C, Dupeux, F, Pica, A, Granier, S, Marquez, J.A. | | 登録日 | 2020-05-01 | | 公開日 | 2021-05-12 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | An automated platform for structural analysis of membrane proteins through serial crystallography.
Cell Rep Methods, 1, 2021
|
|
1L23
 
 | |
