4XBU
 
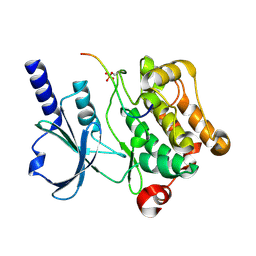 | | In vitro Crystal Structure of PAK4 in complex with Inka peptide | | 分子名称: | Protein FAM212A, Serine/threonine-protein kinase PAK 4 | | 著者 | Baskaran, Y, Ang, K.C, Anekal, P.V, Chan, W.L, Grimes, J.M, Manser, E, Robinson, R.C. | | 登録日 | 2014-12-17 | | 公開日 | 2015-12-02 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.06 Å) | | 主引用文献 | An in cellulo-derived structure of PAK4 in complex with its inhibitor Inka1
Nat Commun, 6, 2015
|
|
4XGY
 
 | | GFP based antibody (fluorobody) | | 分子名称: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, Green fluorescent protein, mAb LCDR3 | | 著者 | Shi, N, Chen, Y.G, Wang, S.H. | | 登録日 | 2015-01-03 | | 公開日 | 2015-07-01 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.494 Å) | | 主引用文献 | The structure of a GFP-based antibody (fluorobody) to TLH, a toxin from Vibrio parahaemolyticus.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
4XL5
 
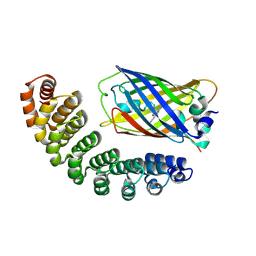 | | X-ray structure of bGFP-A / EGFP complex | | 分子名称: | Green fluorescent protein, bGFP-A | | 著者 | Chevrel, A, Urvoas, A, Li de la Sierra-Gallay, I, Van Tilbeurgh, H, Minard, P, Valerio-Lepiniec, M. | | 登録日 | 2015-01-13 | | 公開日 | 2015-08-19 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Specific GFP-binding artificial proteins ( alpha Rep): a new tool for in vitro to live cell applications.
Biosci.Rep., 35, 2015
|
|
4XOV
 
 | |
4XOW
 
 | |
4XVP
 
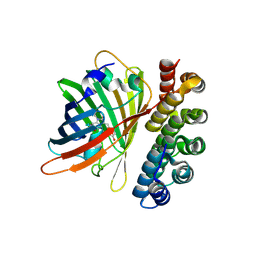 | | X-ray structure of bGFP-C / EGFP complex | | 分子名称: | BGFP-C, Green fluorescent protein | | 著者 | Chevrel, A, Urvoas, A, Li de la Sierra-Gallay, I, Van Tilbeurgh, H, Minard, P, Valerio-Lepiniec, M. | | 登録日 | 2015-01-27 | | 公開日 | 2015-08-19 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | Specific GFP-binding artificial proteins ( alpha Rep): a new tool for in vitro to live cell applications.
Biosci.Rep., 35, 2015
|
|
4Z4K
 
 | |
4Z4M
 
 | |
4ZBL
 
 | |
4ZF3
 
 | |
4ZF4
 
 | |
4ZF5
 
 | |
4ZFS
 
 | |
4ZGY
 
 | |
4ZUZ
 
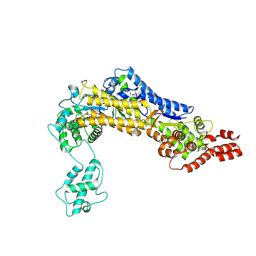 | | SidC 1-871 | | 分子名称: | SidC | | 著者 | Luo, X, Wasilko, D.J, Liu, Y, Sun, J, Wu, X, Luo, Z.-Q, Mao, Y. | | 登録日 | 2015-05-18 | | 公開日 | 2015-07-29 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.86 Å) | | 主引用文献 | Structure of the Legionella Virulence Factor, SidC Reveals a Unique PI(4)P-Specific Binding Domain Essential for Its Targeting to the Bacterial Phagosome.
Plos Pathog., 11, 2015
|
|
4ZVO
 
 | |
4ZVP
 
 | |
4ZVQ
 
 | |
4ZVR
 
 | |
4ZVS
 
 | |
4ZVT
 
 | |
4ZVU
 
 | |
5A2N
 
 | |
5A2O
 
 | |
5A2Q
 
 | | Structure of the HCV IRES bound to the human ribosome | | 分子名称: | 18S RRNA, HCV IRES, MAGNESIUM ION, ... | | 著者 | Quade, N, Leiundgut, M, Boehringer, D, Heuvel, J.v.d, Ban, N. | | 登録日 | 2015-05-21 | | 公開日 | 2015-07-15 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Cryo-Em Structure of Hepatitis C Virus Ires Bound to the Human Ribosome at 3.9 Angstrom Resolution
Nat.Commun., 6, 2015
|
|
