5KP4
 
 | | Crystal Structure of Ketosteroid Isomerase from Pseudomonas putida (pKSI) bound to 19-nortestosterone | | 分子名称: | (8~{R},9~{S},10~{R},13~{S},14~{S},17~{S})-13-methyl-17-oxidanyl-2,6,7,8,9,10,11,12,14,15,16,17-dodecahydro-1~{H}-cyclop enta[a]phenanthren-3-one, Steroid Delta-isomerase | | 著者 | Wu, Y, Boxer, S.G. | | 登録日 | 2016-07-01 | | 公開日 | 2016-09-07 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.706 Å) | | 主引用文献 | A Critical Test of the Electrostatic Contribution to Catalysis with Noncanonical Amino Acids in Ketosteroid Isomerase.
J.Am.Chem.Soc., 138, 2016
|
|
5KP1
 
 | |
5KP3
 
 | |
7U5Z
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with JLJ353 | | 分子名称: | 2-chloro-4-({5-[(2,6-difluorophenyl)methyl]-1,3-oxazol-2-yl}amino)benzonitrile, Reverse transcriptase/ribonuclease H, p51 RT | | 著者 | Hollander, K, Carter, Z, Jorgensen, W.L, Anderson, K.S. | | 登録日 | 2022-03-03 | | 公開日 | 2023-03-15 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Design, synthesis, and biological testing of biphenylmethyloxazole inhibitors targeting HIV-1 reverse transcriptase.
Bioorg.Med.Chem.Lett., 84, 2023
|
|
6Z0O
 
 | | Structure of Affimer-NP bound to Crimean-Congo Haemorrhagic Fever Virus Nucleocapsid Protein | | 分子名称: | Affimer-NP, Nucleocapsid | | 著者 | Alvarez-Rodriguez, B, Tiede, C, Trinh, C, Tomlinson, D, Edwards, T.A, Barr, J.N. | | 登録日 | 2020-05-10 | | 公開日 | 2020-05-27 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.5974 Å) | | 主引用文献 | Characterization and applications of a Crimean-Congo hemorrhagic fever virus nucleoprotein-specific Affimer: Inhibitory effects in viral replication and development of colorimetric diagnostic tests.
Plos Negl Trop Dis, 14, 2020
|
|
1LGU
 
 | | T4 Lysozyme Mutant L99A/M102Q | | 分子名称: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Lysozyme | | 著者 | Wei, B.Q, Baase, W.A, Weaver, L.H, Matthews, B.W, Shoichet, B.K. | | 登録日 | 2002-04-16 | | 公開日 | 2002-05-08 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | A Model Binding Site for Testing Scoring Functions in Molecular Docking
J.Mol.Biol., 322, 2002
|
|
1LGW
 
 | | T4 Lysozyme Mutant L99A/M102Q Bound by 2-fluoroaniline | | 分子名称: | 2-FLUOROANILINE, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | 著者 | Wei, B.Q, Baase, W.A, Weaver, L.H, Matthews, B.W, Shoichet, B.K. | | 登録日 | 2002-04-16 | | 公開日 | 2002-05-08 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | A Model Binding Site for Testing Scoring Functions in Molecular Docking
J.Mol.Biol., 322, 2002
|
|
1LGX
 
 | | T4 Lysozyme Mutant L99A/M102Q Bound by 3,5-difluoroaniline | | 分子名称: | 3,5-DIFLUOROANILINE, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | 著者 | Wei, B.Q, Baase, W.A, Weaver, L.H, Matthews, B.W, Shoichet, B.K. | | 登録日 | 2002-04-16 | | 公開日 | 2002-05-08 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | A Model Binding Site for Testing Scoring Functions in Molecular Docking
J.Mol.Biol., 322, 2002
|
|
1LI3
 
 | | T4 lysozyme mutant L99A/M102Q bound by 3-chlorophenol | | 分子名称: | 3-CHLOROPHENOL, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | 著者 | Wei, B.Q, Baase, W.A, Weaver, L.H, Matthews, B.W, Shoichet, B.K. | | 登録日 | 2002-04-17 | | 公開日 | 2002-05-08 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | A Model Binding Site for Testing Scoring Functions in Molecular Docking
J.Mol.Biol., 322, 2002
|
|
1LI6
 
 | | T4 lysozyme mutant L99A/M102Q bound by 5-methylpyrrole | | 分子名称: | 5-METHYLPYRROLE, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | 著者 | Wei, B.Q, Baase, W.A, Weaver, L.H, Matthews, B.W, Shoichet, B.K. | | 登録日 | 2002-04-17 | | 公開日 | 2002-05-08 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | A Model Binding Site for Testing Scoring Functions in Molecular Docking
J.Mol.Biol., 322, 2002
|
|
1LI2
 
 | | T4 Lysozyme Mutant L99A/M102Q Bound by Phenol | | 分子名称: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Lysozyme, ... | | 著者 | Wei, B.Q, Baase, W.A, Weaver, L.H, Matthews, B.W, Shoichet, B.K. | | 登録日 | 2002-04-17 | | 公開日 | 2002-05-08 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | A Model Binding Site for Testing Scoring Functions in Molecular Docking
J.Mol.Biol., 322, 2002
|
|
6UCN
 
 | | Multi-conformer model of Ketosteroid Isomerase from Pseudomonas Putida (pKSI) bound to Equilenin at 250 K | | 分子名称: | CHLORIDE ION, EQUILENIN, MAGNESIUM ION, ... | | 著者 | Yabukarski, F, Herschlag, D, Biel, J.T, Fraser, J.S. | | 登録日 | 2019-09-16 | | 公開日 | 2020-09-23 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.32 Å) | | 主引用文献 | Assessment of enzyme active site positioning and tests of catalytic mechanisms through X-ray-derived conformational ensembles.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6UCY
 
 | | Multi-conformer model of Ketosteroid Isomerase from Pseudomonas Putida (pKSI) bound to 4-Androstenedione at 250 K | | 分子名称: | 4-ANDROSTENE-3-17-DIONE, CHLORIDE ION, MAGNESIUM ION, ... | | 著者 | Yabukarski, F, Herschlag, D, Biel, J.T, Fraser, J.S. | | 登録日 | 2019-09-18 | | 公開日 | 2020-09-23 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.15 Å) | | 主引用文献 | Assessment of enzyme active site positioning and tests of catalytic mechanisms through X-ray-derived conformational ensembles.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6FU1
 
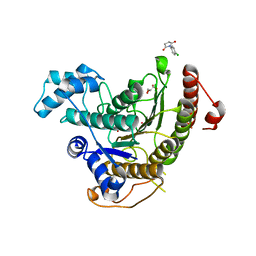 | | Crystal structure of Schistosoma mansoni HDAC8 complexed with a n-alkyl hydroxamate | | 分子名称: | GLYCEROL, Histone deacetylase, POTASSIUM ION, ... | | 著者 | Marek, M, Shaik, T.B, Romier, C. | | 登録日 | 2018-02-26 | | 公開日 | 2019-03-13 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.548 Å) | | 主引用文献 | A Novel Class of Schistosoma mansoni Histone Deacetylase 8 (HDAC8) Inhibitors Identified by Structure-Based Virtual Screening and In Vitro Testing.
Molecules, 23, 2018
|
|
2YGY
 
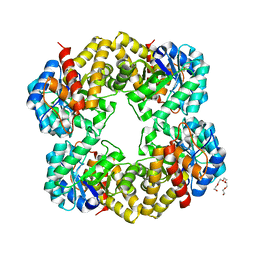 | | Structure of wild type E. coli N-acetylneuraminic acid lyase in space group P21 crystal form II | | 分子名称: | CHLORIDE ION, N-ACETYLNEURAMINATE LYASE, PENTAETHYLENE GLYCOL | | 著者 | Campeotto, I, Nelson, A, Berry, A, Phillips, S.E.V, Pearson, A.R. | | 登録日 | 2011-04-23 | | 公開日 | 2012-04-04 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Pathological macromolecular crystallographic data affected by twinning, partial-disorder and exhibiting multiple lattices for testing of data processing and refinement tools.
Sci Rep, 8, 2018
|
|
3E3A
 
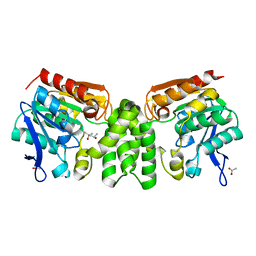 | | The Structure of Rv0554 from Mycobacterium tuberculosis | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, POSSIBLE PEROXIDASE BPOC | | 著者 | Johnston, J.M, Baker, E.N. | | 登録日 | 2008-08-06 | | 公開日 | 2009-06-23 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Structural and functional analysis of Rv0554 from Mycobacterium tuberculosis: testing a putative role in menaquinone biosynthesis.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
1SS1
 
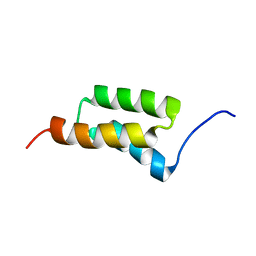 | | STAPHYLOCOCCAL PROTEIN A, B-DOMAIN, Y15W MUTANT, NMR, 25 STRUCTURES | | 分子名称: | Immunoglobulin G binding protein A | | 著者 | Sato, S, Religa, T.L, Daggett, V, Fersht, A.R. | | 登録日 | 2004-03-23 | | 公開日 | 2004-04-06 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | From The Cover: Testing protein-folding simulations by experiment: B domain of protein A.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1MJH
 
 | | Structure-based assignment of the biochemical function of hypothetical protein MJ0577: A test case of structural genomics | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, PROTEIN (ATP-BINDING DOMAIN OF PROTEIN MJ0577) | | 著者 | Zarembinski, T.I, Hung, L.-W, Mueller-Dieckmann, H.J, Kim, K.-K, Yokota, H, Kim, R, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | 登録日 | 1998-11-04 | | 公開日 | 1998-12-23 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structure-based assignment of the biochemical function of a hypothetical protein: a test case of structural genomics.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
6T9S
 
 | | Human Butyrylcholinesterase in complex with 2-(N-hydroxyimino)-N-[(1S)-3-{4-[(2-methyl-1H-imidazol-1-yl)methyl]-1H-1,2,3-triazol-1-yl}-1- phenylpropyl]acetamide | | 分子名称: | (2~{E})-2-hydroxyimino-~{N}-[(1~{S})-3-[4-[(2-methylimidazol-1-yl)methyl]-1,2,3-triazol-1-yl]-1-phenyl-propyl]ethanamid e, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(5-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Brazzolotto, X, Sinko, G, Marakovic, N, Knezevic, A. | | 登録日 | 2019-10-28 | | 公開日 | 2020-07-15 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Enantioseparation, in vitro testing, and structural characterization of triple-binding reactivators of organophosphate-inhibited cholinesterases.
Biochem.J., 477, 2020
|
|
6T9P
 
 | | Human Butyrylcholinesterase in complex with 2-(N-hydroxyimino)-N-[(1R)-3-{4-[(2-methyl-1H-imidazol-1-yl)methyl]-1H-1,2,3-triazol-1-yl}-1- phenylpropyl]acetamide | | 分子名称: | (R,E)-2-(hydroxyimino)-N-(3-(4-((2-methyl-1H-imidazol-1-yl)methyl)-1H-1,2,3-triazol-1-yl)-1-phenylpropyl)acetamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[1-deoxy-alpha-D-tagatopyranose-(2-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Brazzolotto, X, Sinko, G, Marakovic, N, Knezevic, A. | | 登録日 | 2019-10-28 | | 公開日 | 2020-07-15 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Enantioseparation, in vitro testing, and structural characterization of triple-binding reactivators of organophosphate-inhibited cholinesterases.
Biochem.J., 477, 2020
|
|
6UBQ
 
 | |
6UCW
 
 | | Multi-conformer model of Apo Ketosteroid Isomerase from Pseudomonas Putida (pKSI) at 250 K | | 分子名称: | CHLORIDE ION, MAGNESIUM ION, Steroid Delta-isomerase | | 著者 | Yabukarski, F, Herschlag, D, Biel, J.T, Fraser, J.S. | | 登録日 | 2019-09-17 | | 公開日 | 2020-09-23 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Assessment of enzyme active site positioning and tests of catalytic mechanisms through X-ray-derived conformational ensembles.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6U4I
 
 | |
6TZD
 
 | |
6U1Z
 
 | |
