3E1F
 
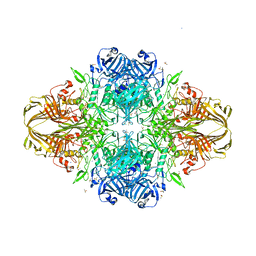 | |
3DYM
 
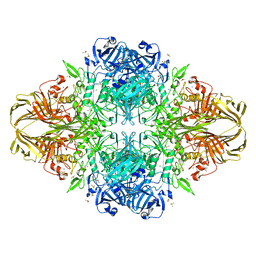 | | E. coli (lacZ) beta-galactosidase (H418E) | | 分子名称: | Beta-galactosidase, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | 著者 | Juers, D.H, Huber, R.E, Matthews, B.W. | | 登録日 | 2008-07-28 | | 公開日 | 2008-10-28 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Direct and indirect roles of His-418 in metal binding and in the activity of beta-galactosidase (E. coli).
Protein Sci., 18, 2009
|
|
3DYP
 
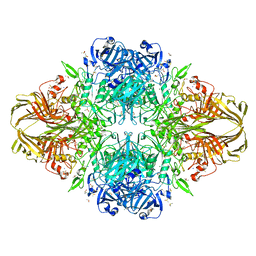 | | E. coli (lacZ) beta-galactosidase (H418N) | | 分子名称: | Beta-galactosidase, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | 著者 | Juers, D.H, Huber, R.E, Matthews, B.W. | | 登録日 | 2008-07-28 | | 公開日 | 2008-10-28 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Direct and indirect roles of His-418 in metal binding and in the activity of beta-galactosidase (E. coli).
Protein Sci., 18, 2009
|
|
4CSY
 
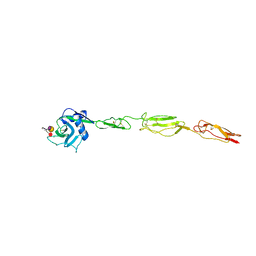 | | E-selectin lectin, EGF-like and two SCR domains complexed with Sialyl Lewis X | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, E-SELECTIN, ... | | 著者 | Preston, R.C, Jakob, R.P, Binder, F.P.C, Sager, C.P, Ernst, B, Maier, T. | | 登録日 | 2014-03-11 | | 公開日 | 2014-09-24 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (2.41 Å) | | 主引用文献 | E-Selectin Ligand Complexes Adopt an Extended High-Affinity Conformation.
J.Mol.Cell.Biol., 8, 2016
|
|
4UIH
 
 | | Cryo-EM structure of Dengue virus serotype 2 strain New Guinea-C complexed with human antibody 2D22 Fab at 37 degree C. The Fab molecules were added to the virus before 37 degree C incubation. | | 分子名称: | ANTIGEN-BINDING FRAGMENT OF HUMAN ANTIBODY 2D22 -HEAVY CHAIN, ANTIGEN-BINDING FRAGMENT OF HUMAN ANTIBODY 2D22 -LIGHT CHAIN, DENGUE VIRUS SEROTYPE 2 STRAIN NEW GUINEA-C E PROTEIN ECTODOMAIN | | 著者 | Fibriansah, G, Ibarra, K.D, Ng, T.-S, Smith, S.A, Tan, J.L, Lim, X.N, Ooi, J.S.G, Kostyuchenko, V.A, Wang, J, de Silva, A.M, Harris, E, Crowe, J.E, Lok, S.-M. | | 登録日 | 2015-03-30 | | 公開日 | 2015-07-15 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (20 Å) | | 主引用文献 | Cryo-EM structure of an antibody that neutralizes dengue virus type 2 by locking E protein dimers.
Science, 349, 2015
|
|
4UMA
 
 | | Structural analysis of substrate-mimicking inhibitors in complex with Neisseria meningitidis 3 deoxy D arabino heptulosonate 7 phosphate synthase the importance of accommodating the active site water | | 分子名称: | (E)-2-METHYL-3-PHOSPHONOACRYLATE, MANGANESE (II) ION, PHOSPHO-2-DEHYDRO-3-DEOXYHEPTONATE ALDOLASE | | 著者 | Heyes, L.C, Reichau, S, Cross, P.J, Parker, E.J. | | 登録日 | 2014-05-16 | | 公開日 | 2014-10-08 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | Structural Analysis of Substrate-Mimicking Inhibitors in Complex with Neisseria Meningitidis 3-Deoxy-D-Arabino-Heptulosonate 7-Phosphate Synthase - the Importance of Accommodating the Active Site Water.
Bioorg.Chem., 57, 2014
|
|
1ED3
 
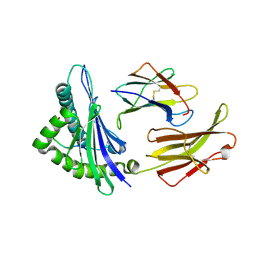 | | CRYSTAL STRUCTURE OF RAT MINOR HISTOCOMPATIBILITY ANTIGEN COMPLEX RT1-AA/MTF-E. | | 分子名称: | BETA-2-MICROGLOBULIN, CLASS I MAJOR HISTOCOMPATIBILITY ANTIGEN RT1-AA, PEPTIDE MTF-E (13N3E) | | 著者 | Speir, J.A, Stevens, J, Joly, E, Butcher, G.W, Wilson, I.A. | | 登録日 | 2000-01-26 | | 公開日 | 2001-02-28 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Two different, highly exposed, bulged structures for an unusually long peptide bound to rat MHC class I RT1-Aa.
Immunity, 14, 2001
|
|
7BH8
 
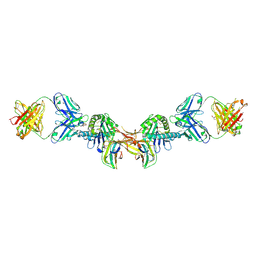 | | 3H4-Fab HLA-E-VL9 co-complex | | 分子名称: | 3H4 Fab heavy chain, 3H4 Fab light chain, Beta-2-microglobulin, ... | | 著者 | Walters, L.C, Rozbesky, D. | | 登録日 | 2021-01-10 | | 公開日 | 2022-04-13 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Mouse and human antibodies bind HLA-E-leader peptide complexes and enhance NK cell cytotoxicity.
Commun Biol, 5, 2022
|
|
8V5P
 
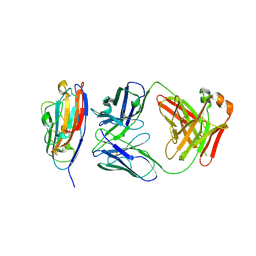 | |
8V5L
 
 | | Structure of the Varicella Zoster Virus (VZV) gI binding domain of glycoprotein E (gE) in complex with human Fab 1A2 and 1E12 | | 分子名称: | Envelope glycoprotein E, Fab 1A2 Heavy Chain, Fab 1A2 Light Chain, ... | | 著者 | Seraj, N, Holzapfel, G, Harshbarger, W. | | 登録日 | 2023-11-30 | | 公開日 | 2024-10-09 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (3.09 Å) | | 主引用文献 | Structures of the Varicella Zoster Virus Glycoprotein E and Epitope Mapping of Vaccine-Elicited Antibodies.
Vaccines (Basel), 12, 2024
|
|
8V5Q
 
 | | Varicella Zoster Virus (VZV) glycoprotein E (gE) gI binding domain in complex with human Fab 1E3 | | 分子名称: | DI(HYDROXYETHYL)ETHER, Envelope glycoprotein E, Fab 1E3 Heavy Chain, ... | | 著者 | Holzapfel, G, Seraj, N, Harshbarger, W. | | 登録日 | 2023-11-30 | | 公開日 | 2024-12-11 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structures of the Varicella Zoster Virus Glycoprotein E and Epitope Mapping of Vaccine-Elicited Antibodies.
Vaccines (Basel), 12, 2024
|
|
7P4B
 
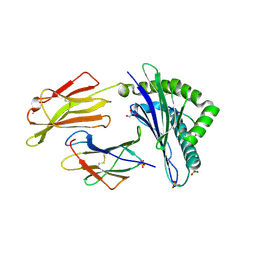 | | HLA-E*01:03 in complex with IL9 | | 分子名称: | Beta-2-microglobulin, ESAT-6-like protein EsxH, GLYCEROL, ... | | 著者 | Walters, L.C, Gillespie, G.M. | | 登録日 | 2021-07-10 | | 公開日 | 2022-07-27 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (1.72 Å) | | 主引用文献 | Primary and secondary functions of HLA-E are determined by stability and conformation of the peptide-bound complexes.
Cell Rep, 39, 2022
|
|
7P49
 
 | | HLA-E*01:03 in complex with Mtb14 | | 分子名称: | Beta-2-microglobulin, GLYCEROL, HLA class I histocompatibility antigen, ... | | 著者 | Walters, L.C, Gillespie, G.M. | | 登録日 | 2021-07-10 | | 公開日 | 2022-06-22 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Primary and secondary functions of HLA-E are determined by stability and conformation of the peptide-bound complexes.
Cell Rep, 39, 2022
|
|
7QB2
 
 | | Pim1 in complex with (E)-4-((6-amino-1-methyl-2-oxoindolin-3-ylidene)methyl)benzoic acid and Pimtide | | 分子名称: | 4-[(E)-(6-azanyl-1-methyl-2-oxidanylidene-indol-3-ylidene)methyl]benzoic acid, GLYCEROL, Pimtide, ... | | 著者 | Hochban, P.M.M, Heine, A, Diederich, W.E. | | 登録日 | 2021-11-18 | | 公開日 | 2022-11-30 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (2.53 Å) | | 主引用文献 | Pose, duplicate, then elaborate: Steps towards increased affinity for inhibitors targeting the specificity surface of the Pim-1 kinase.
Eur.J.Med.Chem., 245, 2023
|
|
6GGM
 
 | | HLA-E*01:03 in complex with the Mtb44 peptide variant: Mtb44*P2-Phe. | | 分子名称: | Beta-2-microglobulin, MHC class I antigen, Mtb44*P2-Phe peptide variant (ARG-PHE-PRO-ALA-LYS-ALA-PRO-LEU-LEU), ... | | 著者 | Walters, L.C, Gillespie, G.M, McMichael, A.J, Rozbesky, D, Jones, E.Y, Harlos, K. | | 登録日 | 2018-05-03 | | 公開日 | 2018-08-08 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.734 Å) | | 主引用文献 | Pathogen-derived HLA-E bound epitopes reveal broad primary anchor pocket tolerability and conformationally malleable peptide binding.
Nat Commun, 9, 2018
|
|
6GH4
 
 | | HLA-E*01:03 in complex with the Mtb44 peptide variant: Mtb44*P2-Gln. | | 分子名称: | ARG-GLN-PRO-ALA-LYS-ALA-PRO-LEU-LEU, Beta-2-microglobulin, MHC class I antigen, ... | | 著者 | Walters, L.C, Gillespie, G.M, McMichael, A.J, Rozbesky, D, Jones, E.Y, Harlos, K. | | 登録日 | 2018-05-04 | | 公開日 | 2018-08-08 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.16 Å) | | 主引用文献 | Pathogen-derived HLA-E bound epitopes reveal broad primary anchor pocket tolerability and conformationally malleable peptide binding.
Nat Commun, 9, 2018
|
|
6GH1
 
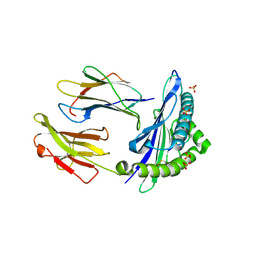 | | HLA-E*01:03 in complex with Mtb44 | | 分子名称: | Beta-2-microglobulin, Enoyl-[acyl-carrier-protein] reductase [NADH], MHC class I antigen, ... | | 著者 | Walters, L.C, Gillespie, G.M, McMichael, A.J, Rozbesky, D, Jones, E.Y, Harlos, K. | | 登録日 | 2018-05-04 | | 公開日 | 2018-08-08 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Pathogen-derived HLA-E bound epitopes reveal broad primary anchor pocket tolerability and conformationally malleable peptide binding.
Nat Commun, 9, 2018
|
|
6GL1
 
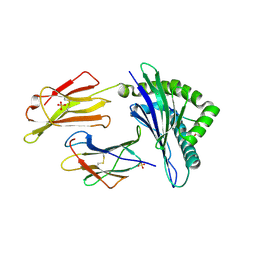 | | HLA-E*01:03 in complex with the HIV epitope, RL9HIV | | 分子名称: | ARG-MET-TYR-SER-PRO-THR-SER-ILE-LEU, Beta-2-microglobulin, MHC class I antigen, ... | | 著者 | Walters, L.C, Gillespie, G.M, McMichael, A.J, Rozbesky, D, Jones, E.Y, Harlos, K. | | 登録日 | 2018-05-22 | | 公開日 | 2018-08-08 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.623 Å) | | 主引用文献 | Pathogen-derived HLA-E bound epitopes reveal broad primary anchor pocket tolerability and conformationally malleable peptide binding.
Nat Commun, 9, 2018
|
|
5KSR
 
 | | Stationary phase survival protein E (SurE) from Xylella fastidiosa - XFSurE-TB (Tetramer Bigger). | | 分子名称: | 5'-nucleotidase SurE, CHLORIDE ION, IODIDE ION, ... | | 著者 | Machado, A.T.P, Fonseca, E.M.B, Dos Reis, M.A, Saraiva, A.M, Dos Santos, C.A, De Toledo, M.A, Polikarpov, I, De Souza, A.P, De Aparicio, R, Iulek, J. | | 登録日 | 2016-07-09 | | 公開日 | 2017-07-19 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | Conformational variability of the stationary phase survival protein E from Xylella fastidiosa revealed by X-ray crystallography, small-angle X-ray scattering studies, and normal mode analysis.
Proteins, 85, 2017
|
|
5KSQ
 
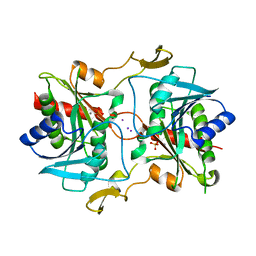 | | Stationary phase survival protein E (SurE) from Xylella fastidiosa | | 分子名称: | 5'-nucleotidase SurE, IODIDE ION, MANGANESE (II) ION, ... | | 著者 | Machado, A.T.P, Fonseca, E.M.B, Dos Reis, M.A, Saraiva, A.M, Dos Santos, C.A, De Toledo, M.A, Polikarpov, I, De Souza, A.P, Aparicio, R, Iulek, J. | | 登録日 | 2016-07-09 | | 公開日 | 2017-07-19 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.63 Å) | | 主引用文献 | Conformational variability of the stationary phase survival protein E from Xylella fastidiosa revealed by X-ray crystallography, small-angle X-ray scattering studies, and normal mode analysis.
Proteins, 85, 2017
|
|
8RLU
 
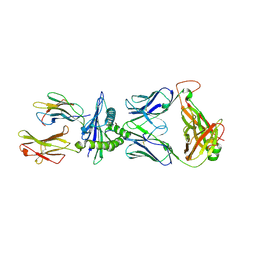 | |
5KSS
 
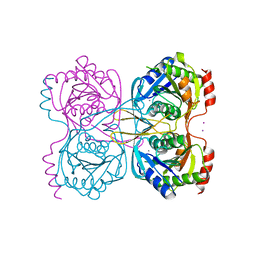 | | Stationary phase survival protein E (SurE) from Xylella fastidiosa - XFSurE-Ds (Dimer Smaller) | | 分子名称: | 5'-nucleotidase SurE, CHLORIDE ION, IODIDE ION, ... | | 著者 | Machado, A.T.P, Fonseca, E.M.B, Dos Reis, M.A, Saraiva, A.M, Dos Santos, C.A, De Toledo, A.M.S, Polikarpov, I, De Souza, A.P, Aparicio, R, Iulek, J. | | 登録日 | 2016-07-09 | | 公開日 | 2017-07-19 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.82 Å) | | 主引用文献 | Conformational variability of the stationary phase survival protein E from Xylella fastidiosa revealed by X-ray crystallography, small-angle X-ray scattering studies, and normal mode analysis.
Proteins, 85, 2017
|
|
5KST
 
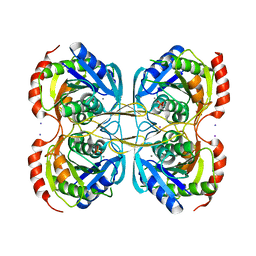 | | Stationary phase Survival protein E (SurE) from Xylella fastidiosa- XfSurE-TSAmp (Tetramer Smaller - crystallization with 3'AMP). | | 分子名称: | 5'-nucleotidase SurE, IODIDE ION, MANGANESE (II) ION, ... | | 著者 | Machado, A.T.P, Fonseca, E.M.B, Dos Reis, M.A, Saraiva, A.M, Dos Santos, C.A, De Toledo, M.A.S, Polikarpov, I, De Souza, A.P, Aparicio, R, Iulek, J. | | 登録日 | 2016-07-09 | | 公開日 | 2017-07-19 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.759 Å) | | 主引用文献 | Conformational variability of the stationary phase survival protein E from Xylella fastidiosa revealed by X-ray crystallography, small-angle X-ray scattering studies, and normal mode analysis.
Proteins, 85, 2017
|
|
9FNS
 
 | |
8RLT
 
 | |
