7Q44
 
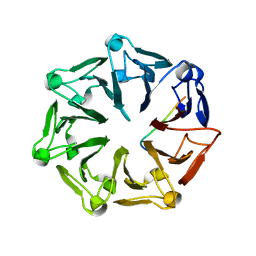 | | Crystal structure of RCC1-Like domain 2 of ubiquitin ligase HERC2 in complex with DXDKDED motif of deubiquitinase USP35 | | 分子名称: | CITRIC ACID, Deubiquitinase USP35 peptide, E3 ubiquitin-protein ligase HERC2 | | 著者 | Demenge, A, Howard, E, Cousido-Siah, A, Mitschler, A, Podjarny, A, McEwen, A.G, Trave, G. | | 登録日 | 2021-10-29 | | 公開日 | 2022-11-16 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.20007777 Å) | | 主引用文献 | Crystal structure of RCC1-Like domain 2 of ubiquitin ligase HERC2 in complex with DXDKDED motif of deubiquitinase USP35
To Be Published
|
|
1XJH
 
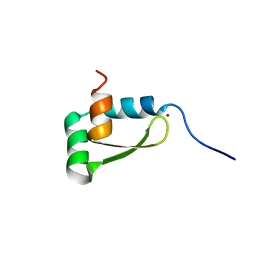 | | NMR structure of the redox switch domain of the E. coli Hsp33 | | 分子名称: | 33 kDa chaperonin, ZINC ION | | 著者 | Won, H.S, Low, L.Y, De Guzman, R.N, Martinez-Yamout, M.A, Jakob, U, Dyson, H.J. | | 登録日 | 2004-09-23 | | 公開日 | 2004-10-05 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The Zinc-dependent Redox Switch Domain of the Chaperone Hsp33 has a Novel Fold
J.Mol.Biol., 341, 2004
|
|
6WEY
 
 | |
1LJ2
 
 | |
2ACF
 
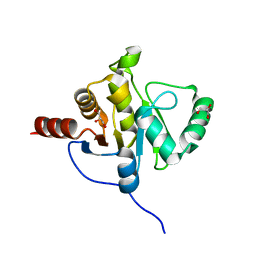 | | NMR STRUCTURE OF SARS-COV NON-STRUCTURAL PROTEIN NSP3A (SARS1) FROM SARS CORONAVIRUS | | 分子名称: | GLYCEROL, Replicase polyprotein 1ab | | 著者 | Saikatendu, K.S, Joseph, J.S, Subramanian, V, Neuman, B.W, Buchmeier, M.J, Stevens, R.C, Kuhn, P, Joint Center for Structural Genomics (JCSG) | | 登録日 | 2005-07-18 | | 公開日 | 2006-02-14 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structural basis of severe acute respiratory syndrome coronavirus ADP-ribose-1''-phosphate dephosphorylation by a conserved domain of nsP3.
Structure, 13, 2005
|
|
8SH8
 
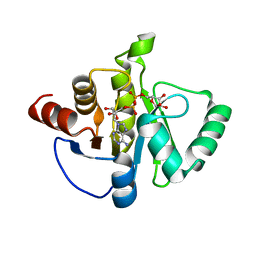 | | Crystal structure of SARS-CoV-2 NSP3 macrodomain Asn40Asp mutant in complex with ADP-ribose (P43 crystal form) | | 分子名称: | Papain-like protease nsp3, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | 著者 | Correy, G.J, Fraser, J.S. | | 登録日 | 2023-04-13 | | 公開日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (0.9 Å) | | 主引用文献 | Crystal structure of SARS-CoV-2 NSP3 macrodomain Asn40Asp mutant in complex with ADP-ribose (P43 crystal form)
To Be Published
|
|
8SH6
 
 | |
1I7F
 
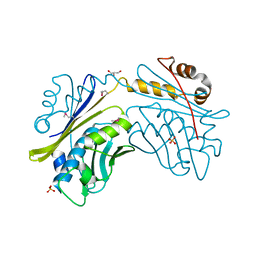 | | CRYSTAL STRUCTURE OF THE HSP33 DOMAIN WITH CONSTITUTIVE CHAPERONE ACTIVITY | | 分子名称: | GLYCEROL, HEAT SHOCK PROTEIN 33, SULFATE ION | | 著者 | Kim, S.-J, Jeong, D.-G, Chi, S.-W, Lee, J.-S, Ryu, S.-E. | | 登録日 | 2001-03-09 | | 公開日 | 2001-05-09 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Crystal structure of proteolytic fragments of the redox-sensitive Hsp33 with constitutive chaperone activity
Nat.Struct.Biol., 8, 2001
|
|
3U12
 
 | | The pleckstrin homology (PH) domain of USP37 | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, SULFATE ION, ... | | 著者 | Dong, A, Nair, U.B, Wernimont, A, Walker, J.R, Weigelt, J, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | 登録日 | 2011-09-29 | | 公開日 | 2011-11-09 | | 最終更新日 | 2012-05-02 | | 実験手法 | X-RAY DIFFRACTION (2.08 Å) | | 主引用文献 | The pleckstrin homology (PH) domain of USP37
To be Published
|
|
6NW9
 
 | | CRYSTAL STRUCTURE OF A TAILSPIKE PROTEIN 3 (TSP3, ORF212) FROM ESCHERICHIA COLI O157:H7 BACTERIOPHAGE CBA120 | | 分子名称: | 1,2-ETHANEDIOL, CARBONATE ION, CHLORIDE ION, ... | | 著者 | Greenfield, J.Y, Herzberg, O. | | 登録日 | 2019-02-06 | | 公開日 | 2019-06-05 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structure and tailspike glycosidase machinery of ORF212 from E. coli O157:H7 phage CBA120 (TSP3).
Sci Rep, 9, 2019
|
|
1PV2
 
 | |
1KNZ
 
 | | Recognition of the rotavirus mRNA 3' consensus by an asymmetric NSP3 homodimer | | 分子名称: | 5'-R(*UP*GP*AP*CP*C)-3', Nonstructural RNA-binding Protein 34 | | 著者 | Deo, R.C, Groft, C.M, Rajashankar, K.R, Burley, S.K. | | 登録日 | 2001-12-19 | | 公開日 | 2002-01-17 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Recognition of the rotavirus mRNA 3' consensus by an asymmetric NSP3 homodimer.
Cell(Cambridge,Mass.), 108, 2002
|
|
8ILC
 
 | |
5OHN
 
 | |
7T9W
 
 | | Crystal structure of the Nsp3 bSM (Betacoronavirus-Specific Marker) domain from SARS-CoV-2 | | 分子名称: | CHLORIDE ION, GLYCEROL, Papain-like protease nsp3 | | 著者 | Stogios, P.J, Skarina, T, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2021-12-20 | | 公開日 | 2021-12-29 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure of the Nsp3 bSM (Betacoronavirus-Specific Marker) domain from SARS-CoV-2
To Be Published
|
|
7THH
 
 | | SUD-C and Ubl2 domains of SARS CoV-2 Nsp3 protein | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, HEXAETHYLENE GLYCOL, ... | | 著者 | Osipiuk, J, Jedrzejczak, R, Endres, M, Wydorski, P, Joachimiak, L, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2022-01-11 | | 公開日 | 2022-01-19 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.32 Å) | | 主引用文献 | SUD-C and Ubl2 domains of SARS CoV-2 Nsp3 protein
to be published
|
|
7TI9
 
 | | Crystal structure of the ubiquitin-like domain 1 (Ubl1) of Nsp3 from SARS-CoV-2, form 2 | | 分子名称: | CHLORIDE ION, GLYCEROL, Papain-like protease nsp3 | | 著者 | Stogios, P.J, Skarina, T, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2022-01-13 | | 公開日 | 2022-01-26 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.73 Å) | | 主引用文献 | Crystal structure of the ubiquitin-like domain 1 (Ubl1) of Nsp3 from SARS-CoV-2, form 2
To Be Published
|
|
7TWR
 
 | |
7TWF
 
 | |
7TWP
 
 | |
7TWQ
 
 | |
7TWG
 
 | |
7TWS
 
 | |
7TWI
 
 | |
7TWJ
 
 | |
