5VLC
 
 | |
6PV6
 
 | | Functional Pathways of Biomolecules Retrieved from Single-particle Snapshots | | 分子名称: | CALCIUM ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ... | | 著者 | Dashti, A, des Georges, A, Frank, J, Ourmazd, A. | | 登録日 | 2019-07-19 | | 公開日 | 2020-08-12 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | Retrieving functional pathways of biomolecules from single-particle snapshots.
Nat Commun, 11, 2020
|
|
1S5T
 
 | |
5VLD
 
 | |
1S5V
 
 | |
5VLB
 
 | |
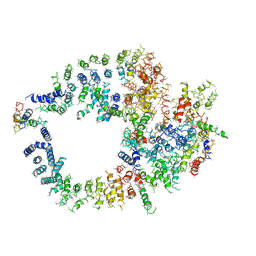
3KGV
 
 | |
6JSH
 
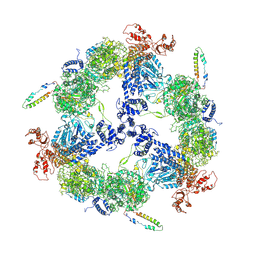 | | Apo-state Fatty Acid Synthase | | 分子名称: | Fatty acid synthase subunit alpha, Fatty acid synthase subunit beta | | 著者 | Qiu, S.W, Liu, S. | | 登録日 | 2019-04-08 | | 公開日 | 2019-09-25 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (5.1 Å) | | 主引用文献 | Modulation of fatty acid synthase by ATR checkpoint kinase Rad3.
J Mol Cell Biol, 11, 2019
|
|
6JSI
 
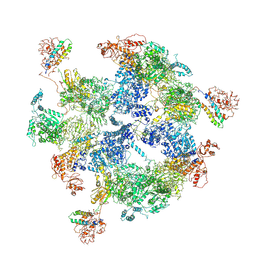 | | Co-purified Fatty Acid Synthase | | 分子名称: | Fatty acid synthase subunit alpha, Fatty acid synthase subunit beta | | 著者 | Qiu, S.W, Liu, S. | | 登録日 | 2019-04-08 | | 公開日 | 2019-09-25 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (4.7 Å) | | 主引用文献 | Modulation of fatty acid synthase by ATR checkpoint kinase Rad3.
J Mol Cell Biol, 11, 2019
|
|
6ND4
 
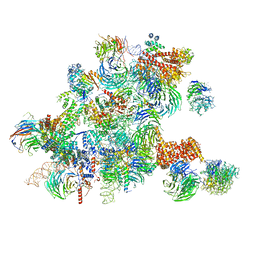 | | Conformational switches control early maturation of the eukaryotic small ribosomal subunit | | 分子名称: | 18S rRNA 5' domain start, 5'ETS rRNA, Bud21, ... | | 著者 | Hunziker, M, Barandun, J, Klinge, S. | | 登録日 | 2018-12-13 | | 公開日 | 2019-06-19 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | Conformational switches control early maturation of the eukaryotic small ribosomal subunit.
Elife, 8, 2019
|
|
8CEC
 
 | | Rnase R bound to a 30S degradation intermediate (State I - head-turning) | | 分子名称: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | 著者 | Paternoga, H, Dimitrova-Paternoga, L, Wilson, D.N. | | 登録日 | 2023-02-01 | | 公開日 | 2023-12-20 | | 最終更新日 | 2024-10-23 | | 実験手法 | ELECTRON MICROSCOPY (3.57 Å) | | 主引用文献 | Structural basis of ribosomal 30S subunit degradation by RNase R.
Nature, 626, 2024
|
|
8CEE
 
 | | Rnase R bound to a 30S degradation intermediate (State I - head-turning) | | 分子名称: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | 著者 | Paternoga, H, Dimitrova-Paternoga, L, Wilson, D.N. | | 登録日 | 2023-02-01 | | 公開日 | 2023-12-20 | | 最終更新日 | 2024-10-23 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structural basis of ribosomal 30S subunit degradation by RNase R.
Nature, 626, 2024
|
|
8CDV
 
 | | Rnase R bound to a 30S degradation intermediate (state II) | | 分子名称: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | 著者 | Paternoga, H, Dimitrova-Paternoga, L, Wilson, D.N. | | 登録日 | 2023-02-01 | | 公開日 | 2023-12-20 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (4.73 Å) | | 主引用文献 | Structural basis of ribosomal 30S subunit degradation by RNase R.
Nature, 626, 2024
|
|
8CDU
 
 | | Rnase R bound to a 30S degradation intermediate (main state) | | 分子名称: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | 著者 | Paternoga, H, Dimitrova-Paternoga, L, Wilson, D.N. | | 登録日 | 2023-02-01 | | 公開日 | 2023-12-20 | | 最終更新日 | 2024-11-13 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structural basis of ribosomal 30S subunit degradation by RNase R.
Nature, 626, 2024
|
|
8CED
 
 | | Rnase R bound to a 30S degradation intermediate (State I - head-turning) | | 分子名称: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | 著者 | Paternoga, H, Dimitrova-Paternoga, L, Wilson, D.N. | | 登録日 | 2023-02-01 | | 公開日 | 2023-12-20 | | 最終更新日 | 2024-11-13 | | 実験手法 | ELECTRON MICROSCOPY (4.15 Å) | | 主引用文献 | Structural basis of ribosomal 30S subunit degradation by RNase R.
Nature, 626, 2024
|
|
5UK5
 
 | |
8QPP
 
 | | Bacillus subtilis MutS2-collided disome complex (stalled 70S) | | 分子名称: | 16S rRNA (1533-MER), 23S ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Park, E, Mackens-Kiani, T, Berhane, R, Esser, H, Erdenebat, C, Burroughs, A.M, Berninghausen, O, Aravind, L, Beckmann, R, Green, R, Buskirk, A.R. | | 登録日 | 2023-10-02 | | 公開日 | 2023-12-27 | | 最終更新日 | 2024-10-23 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | B. subtilis MutS2 splits stalled ribosomes into subunits without mRNA cleavage.
Embo J., 43, 2024
|
|
8QCQ
 
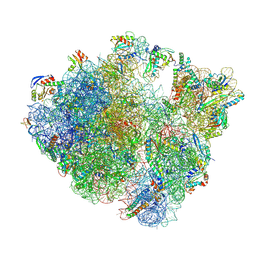 | | B. subtilis ApdA-stalled ribosomal complex | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | 著者 | Morici, M, Wilson, D.N. | | 登録日 | 2023-08-28 | | 公開日 | 2024-03-20 | | 最終更新日 | 2024-11-13 | | 実験手法 | ELECTRON MICROSCOPY (2.3 Å) | | 主引用文献 | RAPP-containing arrest peptides induce translational stalling by short circuiting the ribosomal peptidyltransferase activity.
Nat Commun, 15, 2024
|
|
8R55
 
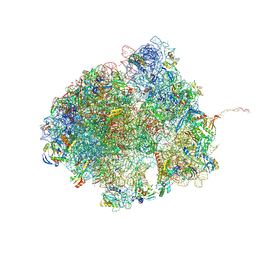 | | Bacillus subtilis MutS2-collided disome complex (collided 70S) | | 分子名称: | 16S rRNA (1533-MER), 23S RNA (2887-MER), 30S ribosomal protein S10, ... | | 著者 | Park, E, Mackens-Kiani, T, Berhane, R, Esser, H, Erdenebat, C, Burroughs, A.M, Berninghausen, O, Aravind, L, Beckmann, R, Green, R, Buskirk, A.R. | | 登録日 | 2023-11-16 | | 公開日 | 2024-01-17 | | 最終更新日 | 2024-10-23 | | 実験手法 | ELECTRON MICROSCOPY (3.57 Å) | | 主引用文献 | B. subtilis MutS2 splits stalled ribosomes into subunits without mRNA cleavage.
Embo J., 43, 2024
|
|
1PAA
 
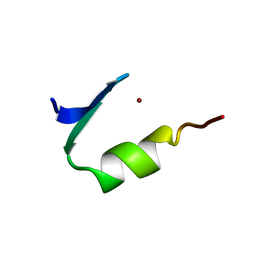 | | STRUCTURE OF A HISTIDINE-X4-HISTIDINE ZINC FINGER DOMAIN: INSIGHTS INTO ADR1-UAS1 PROTEIN-DNA RECOGNITION | | 分子名称: | YEAST TRANSCRIPTION FACTOR ADR1, ZINC ION | | 著者 | Bernstein, B.E, Hoffman, R.C, Horvath, S.J, Herriott, J.R, Klevit, R.E. | | 登録日 | 1994-07-15 | | 公開日 | 1994-10-15 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of a histidine-X4-histidine zinc finger domain: insights into ADR1-UAS1 protein-DNA recognition.
Biochemistry, 33, 1994
|
|
6TPS
 
 | |
6OD1
 
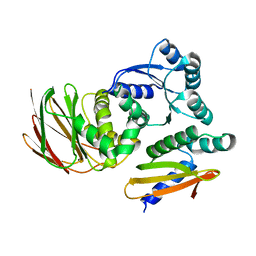 | | IraD-bound to RssB D58P variant | | 分子名称: | Anti-adapter protein IraD, Regulator of RpoS | | 著者 | Deaconescu, A.M, Dorich, V. | | 登録日 | 2019-03-25 | | 公開日 | 2019-04-24 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural basis for inhibition of a response regulator of sigmaSstability by a ClpXP antiadaptor.
Genes Dev., 33, 2019
|
|
6NMI
 
 | | Cryo-EM structure of the human TFIIH core complex | | 分子名称: | CDK-activating kinase assembly factor MAT1, General transcription and DNA repair factor IIH helicase subunit XPB, General transcription and DNA repair factor IIH helicase subunit XPD, ... | | 著者 | Greber, B.J, Toso, D, Fang, J, Nogales, E. | | 登録日 | 2019-01-10 | | 公開日 | 2019-03-13 | | 最終更新日 | 2024-11-06 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | The complete structure of the human TFIIH core complex.
Elife, 8, 2019
|
|
6VJ4
 
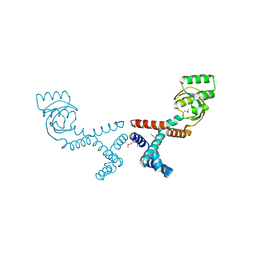 | | 1.70 Angstrom Resolution Crystal Structure of Peptidylprolyl Isomerase (PrsA) from Bacillus anthracis | | 分子名称: | Peptidylprolyl isomerase PrsA | | 著者 | Minasov, G, Shuvalova, L, Kiryukhina, O, Wiersum, G, Endres, M, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-01-14 | | 公開日 | 2020-02-05 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | 1.70 Angstrom Resolution Crystal Structure of Peptidylprolyl Isomerase (PrsA) from Bacillus anthracis
To Be Published
|
|
6Z1P
 
 | |
