6D0Q
 
 | |
1XJX
 
 | | The crystal structures of the DNA binding sites of the RUNX1 transcription factor | | 分子名称: | 5'-D(*TP*CP*TP*GP*CP*GP*GP*TP*C)-3', 5'-D(*TP*GP*AP*CP*CP*GP*CP*AP*G)-3' | | 著者 | Kitayner, M, Rozenberg, H, Rabinovich, D, Shakked, Z. | | 登録日 | 2004-09-26 | | 公開日 | 2005-03-15 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structures of the DNA-binding site of Runt-domain transcription regulators.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
5YYU
 
 | |
6D0R
 
 | |
3KDE
 
 | | Crystal structure of the THAP domain from D. melanogaster P-element transposase in complex with its natural DNA binding site | | 分子名称: | 5'-D(*(BRU)P*CP*CP*AP*CP*TP*TP*AP*AP*C)-3', 5'-D(*GP*TP*TP*AP*AP*GP*(BRU)P*GP*GP*A)-3', Transposable element P transposase, ... | | 著者 | Sabogal, A, Lyubimov, A.Y, Berger, J.M, Rio, D.C. | | 登録日 | 2009-10-22 | | 公開日 | 2009-12-08 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.74 Å) | | 主引用文献 | THAP proteins target specific DNA sites through bipartite recognition of adjacent major and minor grooves.
Nat.Struct.Mol.Biol., 17, 2010
|
|
1F30
 
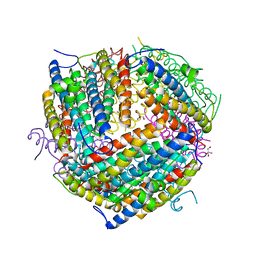 | | THE STRUCTURAL BASIS FOR DNA PROTECTION BY E. COLI DPS PROTEIN | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DNA PROTECTION DURING STARVATION PROTEIN, ZINC ION | | 著者 | Luo, J, Liu, D, White, M.A, Fox, R.O. | | 登録日 | 2000-05-31 | | 公開日 | 2003-06-17 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | The Structural Basis for DNA Protection by E. coli Dps Protein
To be Published
|
|
3SAW
 
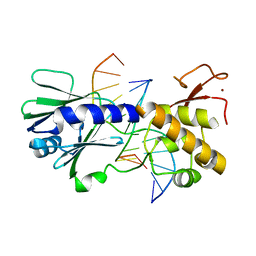 | | MUTM Slanted complex 8 with R112A mutation | | 分子名称: | 5'-D(*A*GP*GP*TP*AP*GP*AP*CP*AP*AP*GP*GP*AP*CP*GP*C)-3', 5'-D(*TP*GP*CP*GP*TP*C*CP*TP*TP*GP*TP*(CX2)P*TP*AP*CP*C)-3', DNA GLYCOSYLASE, ... | | 著者 | Qi, Y, Verdine, G.L. | | 登録日 | 2011-06-03 | | 公開日 | 2012-01-11 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Strandwise translocation of a DNA glycosylase on undamaged DNA.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4NDI
 
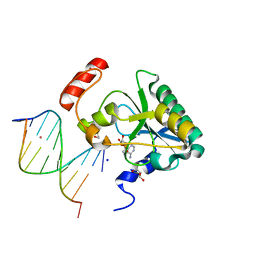 | | Human Aprataxin (Aptx) AOA1 variant K197Q bound to RNA-DNA, AMP, and Zn - product complex | | 分子名称: | 5'-D(*GP*AP*AP*TP*CP*AP*TP*AP*AP*C)-3', 5'-R(P*G)-D(P*TP*TP*AP*TP*GP*AP*TP*TP*C)-3', ADENOSINE MONOPHOSPHATE, ... | | 著者 | Schellenberg, M.J, Tumbale, P.S, Williams, R.S. | | 登録日 | 2013-10-26 | | 公開日 | 2013-12-18 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Aprataxin resolves adenylated RNA-DNA junctions to maintain genome integrity.
Nature, 506, 2013
|
|
4NDF
 
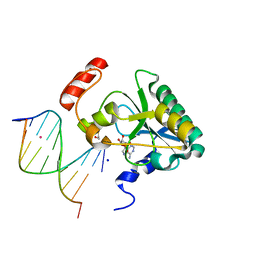 | | Human Aprataxin (Aptx) bound to RNA-DNA, AMP, and Zn - product complex | | 分子名称: | 5'-D(*GP*AP*AP*TP*CP*AP*TP*AP*AP*C)-3', 5'-R(P*G)-D(P*TP*TP*AP*TP*GP*AP*TP*TP*C)-3', ADENOSINE MONOPHOSPHATE, ... | | 著者 | Schellenberg, M.J, Tumbale, P.S, Williams, R.S. | | 登録日 | 2013-10-26 | | 公開日 | 2013-12-18 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.944 Å) | | 主引用文献 | Aprataxin resolves adenylated RNA-DNA junctions to maintain genome integrity.
Nature, 506, 2013
|
|
4NDG
 
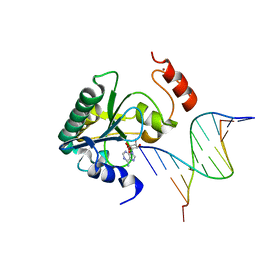 | | Human Aprataxin (Aptx) bound to RNA-DNA and Zn - adenosine vanadate transition state mimic complex | | 分子名称: | 5'-D(*GP*AP*AP*TP*CP*AP*TP*AP*AP*C)-3', 5'-R(P*G)-D(P*TP*TP*AP*TP*GP*AP*TP*TP*C)-3', Aprataxin, ... | | 著者 | Schellenberg, M.J, Tumbale, P.S, Williams, R.S. | | 登録日 | 2013-10-26 | | 公開日 | 2013-12-18 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.541 Å) | | 主引用文献 | Aprataxin resolves adenylated RNA-DNA junctions to maintain genome integrity.
Nature, 506, 2013
|
|
3Q8D
 
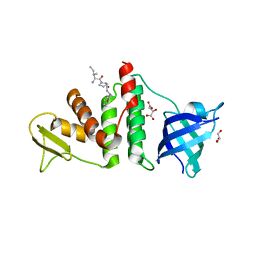 | | E. coli RecO complex with SSB C-terminus | | 分子名称: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, D-MALATE, DNA repair protein recO, ... | | 著者 | Koroleva, O, Ryzhikov, M, Korolev, S. | | 登録日 | 2011-01-06 | | 公開日 | 2011-05-04 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Mechanism of RecO recruitment to DNA by single-stranded DNA binding protein.
Nucleic Acids Res., 39, 2011
|
|
2NNU
 
 | |
4PUO
 
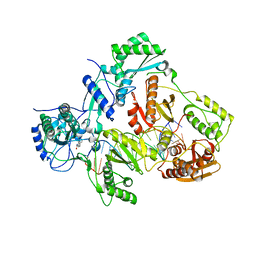 | | Crystal structure of HIV-1 reverse transcriptase in complex with RNA/DNA and Nevirapine | | 分子名称: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, 5'-D(*A*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*GP*CP*GP*CP*CP*G)-3', 5'-R(P*AP*UP*GP*GP*UP*CP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*UP*GP*UP*G)-3', ... | | 著者 | Das, K, Martinez, S.E, Arnold, E. | | 登録日 | 2014-03-13 | | 公開日 | 2014-06-18 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.901 Å) | | 主引用文献 | Structures of HIV-1 RT-RNA/DNA ternary complexes with dATP and nevirapine reveal conformational flexibility of RNA/DNA: insights into requirements for RNase H cleavage.
Nucleic Acids Res., 42, 2014
|
|
4Q0B
 
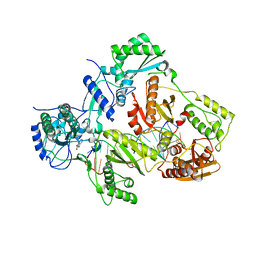 | | Crystal structure of HIV-1 reverse transcriptase in complex with gap-RNA/DNA and Nevirapine | | 分子名称: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, 5'-D(*A*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*GP*CP*GP*CP*CP*G)-3', 5'-R(*AP*UP*GP*GP*UP*CP*GP*GP*CP*GP*CP*CP*CP*G)-3', ... | | 著者 | Das, K, Martinez, S.E, Arnold, E. | | 登録日 | 2014-04-01 | | 公開日 | 2014-06-18 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Structures of HIV-1 RT-RNA/DNA ternary complexes with dATP and nevirapine reveal conformational flexibility of RNA/DNA: insights into requirements for RNase H cleavage.
Nucleic Acids Res., 42, 2014
|
|
6SJF
 
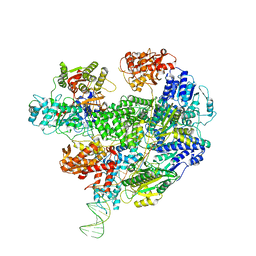 | | Cryo-EM structure of the RecBCD Chi unrecognised complex | | 分子名称: | Forked DNA substrate, RecBCD enzyme subunit RecB, RecBCD enzyme subunit RecC, ... | | 著者 | Cheng, K, Wilkinson, M, Wigley, D.B. | | 登録日 | 2019-08-13 | | 公開日 | 2020-01-01 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | A conformational switch in response to Chi converts RecBCD from phage destruction to DNA repair.
Nat.Struct.Mol.Biol., 27, 2020
|
|
2TBD
 
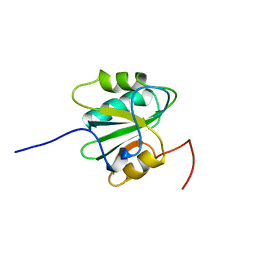 | | SV40 T ANTIGEN DNA-BINDING DOMAIN, NMR, 30 STRUCTURES | | 分子名称: | SV40 T ANTIGEN | | 著者 | Luo, X, Sanford, D.G, Bullock, P.A, Bachovchin, W.W. | | 登録日 | 1997-01-09 | | 公開日 | 1997-04-01 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the origin DNA-binding domain of SV40 T-antigen.
Nat.Struct.Biol., 3, 1996
|
|
6SJG
 
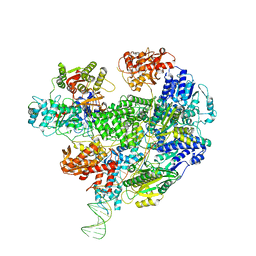 | | Cryo-EM structure of the RecBCD no Chi negative control complex | | 分子名称: | Forked DNA substrate, RecBCD enzyme subunit RecB, RecBCD enzyme subunit RecC, ... | | 著者 | Cheng, K, Wilkinson, M, Wigley, D.B. | | 登録日 | 2019-08-13 | | 公開日 | 2020-01-01 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | A conformational switch in response to Chi converts RecBCD from phage destruction to DNA repair.
Nat.Struct.Mol.Biol., 27, 2020
|
|
4PWD
 
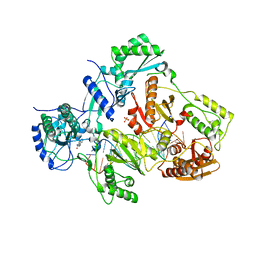 | | Crystal structure of HIV-1 reverse transcriptase in complex with bulge-RNA/DNA and Nevirapine | | 分子名称: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, 5'-D(*A*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*GP*CP*GP*CP*CP*G)-3', 5'-R(*AP*UP*GP*GP*UP*CP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*AP*CP*AP*GP*GP*GP*AP*CP*UP*GP*U)-3', ... | | 著者 | Das, K, Martinez, S.E, Arnold, E. | | 登録日 | 2014-03-19 | | 公開日 | 2014-06-18 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structures of HIV-1 RT-RNA/DNA ternary complexes with dATP and nevirapine reveal conformational flexibility of RNA/DNA: insights into requirements for RNase H cleavage.
Nucleic Acids Res., 42, 2014
|
|
7BTQ
 
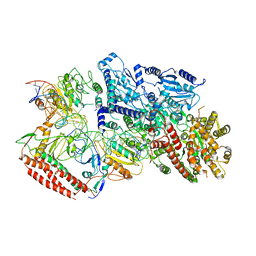 | | EcoR124I-DNA in the Restriction-Alleviation State | | 分子名称: | DNA (64-MER), Type I restriction enzyme EcoR124II M protein, Type I restriction enzyme R Protein, ... | | 著者 | Gao, Y, Gao, P. | | 登録日 | 2020-04-02 | | 公開日 | 2020-05-27 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (4.54 Å) | | 主引用文献 | Structural insights into assembly, operation and inhibition of a type I restriction-modification system.
Nat Microbiol, 5, 2020
|
|
8U8M
 
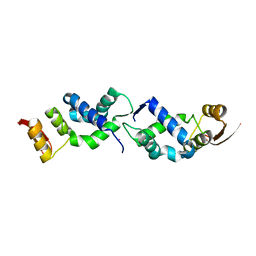 | | X-ray crystal structure of TEBP-1 MCD2 homodimer | | 分子名称: | COBALT (II) ION, Double-strand telomeric DNA-binding proteins 1 | | 著者 | Nandakumar, J, Padmanaban, S. | | 登録日 | 2023-09-18 | | 公開日 | 2024-04-03 | | 最終更新日 | 2024-04-24 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Caenorhabditis elegans telomere-binding proteins TEBP-1 and TEBP-2 adapt the Myb module to dimerize and bind telomeric DNA.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
4PQU
 
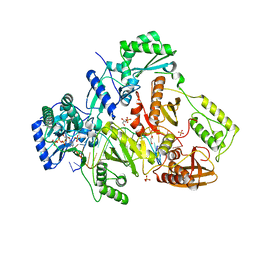 | | Crystal structure of HIV-1 Reverse Transcriptase in complex with RNA/DNA and dATP | | 分子名称: | 1,2-ETHANEDIOL, 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 5'-D(*AP*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*GP*CP*GP*CP*CP*G)-3', ... | | 著者 | Das, K, Bandwar, R.P, Arnold, E. | | 登録日 | 2014-03-04 | | 公開日 | 2014-06-18 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.508 Å) | | 主引用文献 | Structures of HIV-1 RT-RNA/DNA ternary complexes with dATP and nevirapine reveal conformational flexibility of RNA/DNA: insights into requirements for RNase H cleavage.
Nucleic Acids Res., 42, 2014
|
|
6BHJ
 
 | | Structure of HIV-1 Reverse Transcriptase Bound to a 38-mer Hairpin Template-Primer RNA-DNA Aptamer | | 分子名称: | 38-MER RNA-DNA Aptamer, GLYCEROL, HIV-1 REVERSE TRANSCRIPTASE P51 subunit, ... | | 著者 | Ruiz, F.X, Miller, M.T, Tuske, S, Das, K, Arnold, E. | | 登録日 | 2017-10-30 | | 公開日 | 2018-10-31 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.81 Å) | | 主引用文献 | Integrative Structural Biology Studies of HIV-1 Reverse Transcriptase Binding to a High-Affinity DNA Aptamer
Curr Res Struct Biol, 2020
|
|
5WMB
 
 | | Structure of the 10S (-)-cis-BP-dG modified Rev1 ternary complex (the BP residue is disordered) | | 分子名称: | 1,2-ETHANEDIOL, 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Rechkoblit, O, Kolbanovsky, A, Landes, H, Geacintov, N.E, Aggarwal, A.K. | | 登録日 | 2017-07-28 | | 公開日 | 2017-10-25 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Mechanism of error-free replication across benzo[a]pyrene stereoisomers by Rev1 DNA polymerase.
Nat Commun, 8, 2017
|
|
8JZV
 
 | | RPA70N-ETAA1 fusion | | 分子名称: | Ewing's tumor-associated antigen 1, Replication protein A 70 kDa DNA-binding subunit | | 著者 | Fu, W.M, Wu, Y.Y, Zhou, C. | | 登録日 | 2023-07-06 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structural characterization of human RPA70N association with DNA damage response proteins.
Elife, 12, 2023
|
|
8JZY
 
 | | RPA70N-RAD9 fusion | | 分子名称: | Cell cycle checkpoint control protein RAD9A, Replication protein A 70 kDa DNA-binding subunit | | 著者 | Fu, W.M, Wu, Y.Y, Zhou, C. | | 登録日 | 2023-07-06 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structural characterization of human RPA70N association with DNA damage response proteins.
Elife, 12, 2023
|
|
