5IFD
 
 | |
1TUJ
 
 | | Solution structure of the honey bee general odorant binding protein ASP2 in complex with trimethylsilyl-d4 propionate | | 分子名称: | 3-TRIMETHYLSILYL-PROPIONATE-2,2,3,3,-D4, odorant binding protein ASP2 | | 著者 | Lescop, E, Briand, L, Pernollet, J.-C, Guittet, E. | | 登録日 | 2004-06-25 | | 公開日 | 2005-09-20 | | 最終更新日 | 2022-03-02 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the honey bee general odorant binding protein ASP2 in complex with trimethylsilyl-d4 propionate
To be Published
|
|
5IFC
 
 | |
5IF8
 
 | |
5IF7
 
 | |
1AXO
 
 | | STRUCTURAL ALIGNMENT OF THE (+)-TRANS-ANTI-[BP]DG ADDUCT POSITIONED OPPOSITE DC AT A DNA TEMPLATE-PRIMER JUNCTION, NMR, 6 STRUCTURES | | 分子名称: | 1,2,3-TRIHYDROXY-1,2,3,4-TETRAHYDROBENZO[A]PYRENE, DNA DUPLEX D(AAC-[BP]G-CTACCATCC)D(GGATGGTAGC) | | 著者 | Feng, B, Gorin, A.A, Hingerty, B.E, Geacintov, N.E, Broyde, S, Patel, D.J. | | 登録日 | 1997-10-16 | | 公開日 | 1998-07-01 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural alignment of the (+)-trans-anti-benzo[a]pyrene-dG adduct positioned opposite dC at a DNA template-primer junction.
Biochemistry, 36, 1997
|
|
6VAR
 
 | | 61 nt human Hepatitis B virus epsilon pre-genomic RNA | | 分子名称: | RNA (61-MER) | | 著者 | LeBlanc, R.M, Kasprzak, W.K, Longhini, A.P, Abulwerdi, F, Ginocchio, S, Shields, B, Nyman, J, Svirydava, M, Del Vecchio, C, Ivanic, J, Schneekloth, J.S, Dayie, T.K, Shapiro, B.A, Le Grice, S.F.J. | | 登録日 | 2019-12-17 | | 公開日 | 2020-12-30 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR, SOLUTION SCATTERING | | 主引用文献 | Structural insights of the conserved "priming loop" of hepatitis B virus pre-genomic RNA.
J.Biomol.Struct.Dyn., 2021
|
|
1A9H
 
 | |
1E41
 
 | |
1A9G
 
 | |
5IFB
 
 | |
1A9J
 
 | |
6UCK
 
 | | proIAPP in DPC Micelles - Two-Conformer Ensemble Refinement, Bent Conformer | | 分子名称: | Islet amyloid polypeptide | | 著者 | DeLisle, C.F, Malooley, A.L, Banerjee, I, Lorieau, J.L. | | 登録日 | 2019-09-16 | | 公開日 | 2020-02-26 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Pro-islet amyloid polypeptide in micelles contains a helical prohormone segment.
Febs J., 287, 2020
|
|
6UCJ
 
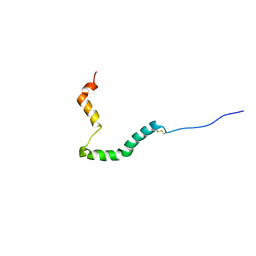 | | proIAPP in DPC Micelles - Two-Conformer Ensemble Refinement, Open Conformer | | 分子名称: | Islet amyloid polypeptide | | 著者 | DeLisle, C.F, Malooley, A.L, Banerjee, I, Lorieau, J.L. | | 登録日 | 2019-09-16 | | 公開日 | 2020-02-26 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Pro-islet amyloid polypeptide in micelles contains a helical prohormone segment.
Febs J., 287, 2020
|
|
6TO6
 
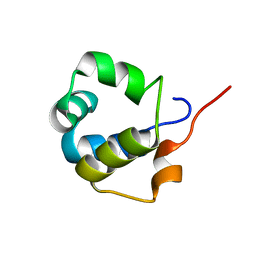 | | Solution structure of the modulator of repression (MOR) of the temperate bacteriophage TP901-1 from Lactococcus lactis | | 分子名称: | MOR | | 著者 | Rasmussen, K.K, Blackledge, M, Herrmann, T, Lo Leggio, L, Jensen, M.R. | | 登録日 | 2019-12-11 | | 公開日 | 2020-08-19 | | 最終更新日 | 2024-06-19 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Revealing the mechanism of repressor inactivation during switching of a temperate bacteriophage.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
193D
 
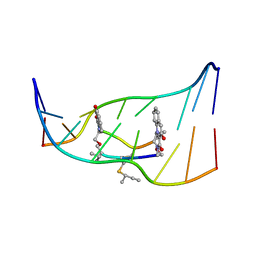 | |
1DK3
 
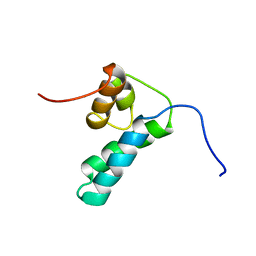 | | REFINED SOLUTION STRUCTURE OF THE N-TERMINAL DOMAIN OF DNA POLYMERASE BETA | | 分子名称: | DNA POLYMERASE BETA | | 著者 | Maciejewski, M.W, Prasad, R, Liu, D.-J, Wilson, S.H, Mullen, G.P. | | 登録日 | 1999-12-06 | | 公開日 | 2000-02-14 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Backbone dynamics and refined solution structure of the N-terminal domain of DNA polymerase beta. Correlation with DNA binding and dRP lyase activity.
J.Mol.Biol., 296, 2000
|
|
1EUB
 
 | | SOLUTION STRUCTURE OF THE CATALYTIC DOMAIN OF HUMAN COLLAGENASE-3 (MMP-13) COMPLEXED TO A POTENT NON-PEPTIDIC SULFONAMIDE INHIBITOR | | 分子名称: | 1-METHYLOXY-4-SULFONE-BENZENE, 3-METHYLPYRIDINE, CALCIUM ION, ... | | 著者 | Zhang, X, Gonnella, N.C, Koehn, J, Pathak, N, Ganu, V, Melton, R, Parker, D, Hu, S.I, Nam, K.Y. | | 登録日 | 2000-04-14 | | 公開日 | 2001-04-14 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the catalytic domain of human collagenase-3 (MMP-13) complexed to a potent non-peptidic sulfonamide inhibitor: binding comparison with stromelysin-1 and collagenase-1.
J.Mol.Biol., 301, 2000
|
|
1A1T
 
 | | STRUCTURE OF THE HIV-1 NUCLEOCAPSID PROTEIN BOUND TO THE SL3 PSI-RNA RECOGNITION ELEMENT, NMR, 25 STRUCTURES | | 分子名称: | NUCLEOCAPSID PROTEIN, SL3 STEM-LOOP RNA, ZINC ION | | 著者 | De Guzman, R.N, Wu, Z.R, Stalling, C.C, Pappalardo, L, Borer, P.N, Summers, M.F. | | 登録日 | 1997-12-15 | | 公開日 | 1998-06-17 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of the HIV-1 nucleocapsid protein bound to the SL3 psi-RNA recognition element.
Science, 279, 1998
|
|
6V6T
 
 | |
1D6X
 
 | |
1D7Q
 
 | | HUMAN TRANSLATION INITIATION FACTOR EIF1A | | 分子名称: | PROTEIN (N-TERMINAL HISTIDINE TAG), TRANSLATION INITIATION FACTOR 1A | | 著者 | Battiste, J.L, Pestova, T.V, Hellen, C.U.T, Wagner, G. | | 登録日 | 1999-10-19 | | 公開日 | 2000-03-17 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The eIF1A solution structure reveals a large RNA-binding surface important for scanning function.
Mol.Cell, 5, 2000
|
|
1E3Y
 
 | |
1DK2
 
 | | REFINED SOLUTION STRUCTURE OF THE N-TERMINAL DOMAIN OF DNA POLYMERASE BETA | | 分子名称: | DNA POLYMERASE BETA | | 著者 | Maciejewski, M.W, Prasad, R, Liu, D.-J, Wilson, S.H, Mullen, G.P. | | 登録日 | 1999-12-06 | | 公開日 | 2000-02-14 | | 最終更新日 | 2024-04-10 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Backbone dynamics and refined solution structure of the N-terminal domain of DNA polymerase beta. Correlation with DNA binding and dRP lyase activity.
J.Mol.Biol., 296, 2000
|
|
1COE
 
 | |
