5JR4
 
 | |
5JVX
 
 | |
3OKF
 
 | | 2.5 Angstrom Resolution Crystal Structure of 3-Dehydroquinate Synthase (aroB) from Vibrio cholerae | | 分子名称: | 3-dehydroquinate synthase, CHLORIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | 著者 | Minasov, G, Light, S.H, Shuvalova, L, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2010-08-24 | | 公開日 | 2010-09-08 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | 2.5 Angstrom Resolution Crystal Structure of 3-Dehydroquinate Synthase (aroB) from Vibrio cholerae.
TO BE PUBLISHED
|
|
6PAB
 
 | |
1EER
 
 | |
5KBM
 
 | | Candida Albicans Superoxide Dismutase 5 (SOD5), D113N Mutant | | 分子名称: | COPPER (I) ION, Cell surface Cu-only superoxide dismutase 5, SULFATE ION | | 著者 | Galaleldeen, A, Peterson, R.L, Villarreal, J, Taylor, A.B, Hart, P.J. | | 登録日 | 2016-06-03 | | 公開日 | 2016-08-24 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (1.416 Å) | | 主引用文献 | The Phylogeny and Active Site Design of Eukaryotic Copper-only Superoxide Dismutases.
J.Biol.Chem., 291, 2016
|
|
5KII
 
 | |
5KIS
 
 | |
5JQ0
 
 | | Crystal structure of human carbonic anhydrase II in complex with Benzoxaborole at pH=8.7 | | 分子名称: | 1,1-dihydroxy-1,3-dihydro-2,1-benzoxaborol-1-ium, Carbonic anhydrase 2, ZINC ION | | 著者 | Alterio, V, Esposito, D, Di Fiore, A, De Simone, G. | | 登録日 | 2016-05-04 | | 公開日 | 2016-10-19 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Benzoxaborole as a new chemotype for carbonic anhydrase inhibition.
Chem.Commun.(Camb.), 52, 2016
|
|
6PAA
 
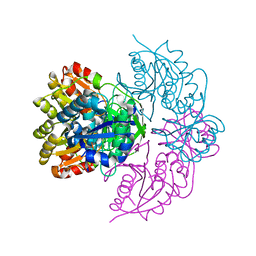 | |
3OT5
 
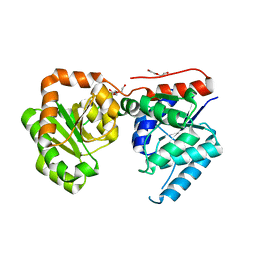 | | 2.2 Angstrom Resolution Crystal Structure of putative UDP-N-acetylglucosamine 2-epimerase from Listeria monocytogenes | | 分子名称: | DI(HYDROXYETHYL)ETHER, TRIETHYLENE GLYCOL, UDP-N-acetylglucosamine 2-epimerase | | 著者 | Minasov, G, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2010-09-10 | | 公開日 | 2010-09-22 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | 2.2 Angstrom Resolution Crystal Structure of putative UDP-N-acetylglucosamine 2-epimerase from Listeria monocytogenes.
TO BE PUBLISHED
|
|
3PAJ
 
 | | 2.00 Angstrom resolution crystal structure of a quinolinate phosphoribosyltransferase from Vibrio cholerae O1 biovar eltor str. N16961 | | 分子名称: | MAGNESIUM ION, Nicotinate-nucleotide pyrophosphorylase, carboxylating | | 著者 | Halavaty, A.S, Minasov, G, Winsor, J, Dubrovska, I, Shuvalova, L, Peterson, S, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2010-10-19 | | 公開日 | 2010-11-03 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | 2.00 Angstrom resolution crystal structure of a quinolinate phosphoribosyltransferase from Vibrio cholerae O1 biovar eltor str. N16961
To be Published
|
|
3OUT
 
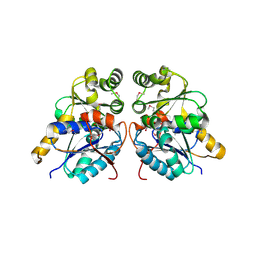 | | Crystal structure of glutamate racemase from Francisella tularensis subsp. tularensis SCHU S4 in complex with D-glutamate. | | 分子名称: | D-GLUTAMIC ACID, Glutamate racemase | | 著者 | Filippova, E.V, Wawrzak, Z, Onopriyenko, O, Kudriska, M, Edwards, A, Savchenko, A, Anderson, F.W, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2010-09-15 | | 公開日 | 2010-09-29 | | 最終更新日 | 2024-11-27 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Crystal structure of glutamate racemase from Francisella tularensis subsp. tularensis SCHU S4 in complex with D-glutamate.
To be Published
|
|
6PA6
 
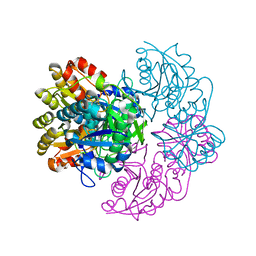 | |
3SGW
 
 | |
6MLJ
 
 | | Crystal structure of the periplasmic Lysine-, Arginine-, Ornithine-binding protein (LAO) S70A mutant from Salmonella typhimurium complexed with arginine | | 分子名称: | ACETATE ION, ARGININE, Lysine/arginine/ornithine transport protein | | 著者 | Romero-Romero, S, Vergara, R, Espinoza-Perez, G, Rodriguez-Romero, A. | | 登録日 | 2018-09-27 | | 公開日 | 2019-08-07 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | The interplay of protein-ligand and water-mediated interactions shape affinity and selectivity in the LAO binding protein.
Febs J., 287, 2020
|
|
6MLV
 
 | | Crystal structure of the periplasmic Lysine-, Arginine-, Ornithine-binding protein (LAO) Y14A mutant from Salmonella typhimurium | | 分子名称: | Lysine/arginine/ornithine-binding periplasmic protein | | 著者 | Romero-Romero, S, Vergara, R, Espinoza-Perez, G, Rodriguez-Romero, A. | | 登録日 | 2018-09-28 | | 公開日 | 2019-08-07 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.082 Å) | | 主引用文献 | The interplay of protein-ligand and water-mediated interactions shape affinity and selectivity in the LAO binding protein.
Febs J., 287, 2020
|
|
6ML0
 
 | | Crystal structure of the periplasmic Lysine-, Arginine-, Ornithine-binding protein (LAO) S69A mutant from Salmonella typhimurium | | 分子名称: | ACETATE ION, GLYCEROL, Lysine/arginine/ornithine-binding periplasmic protein | | 著者 | Romero-Romero, S, Vergara, R, Espinoza-Perez, G, Rodriguez-Romero, A. | | 登録日 | 2018-09-26 | | 公開日 | 2019-08-07 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.68 Å) | | 主引用文献 | The interplay of protein-ligand and water-mediated interactions shape affinity and selectivity in the LAO binding protein.
Febs J., 287, 2020
|
|
6MLO
 
 | | Crystal structure of the periplasmic Lysine-, Arginine-, Ornithine-binding protein (LAO) Y14A mutant from Salmonella typhimurium complexed with arginine | | 分子名称: | ACETATE ION, ARGININE, GLYCEROL, ... | | 著者 | Romero-Romero, S, Vergara, R, Espinoza-Perez, G, Rodriguez-Romero, A. | | 登録日 | 2018-09-27 | | 公開日 | 2019-08-07 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (1.717 Å) | | 主引用文献 | The interplay of protein-ligand and water-mediated interactions shape affinity and selectivity in the LAO binding protein.
Febs J., 287, 2020
|
|
3OPQ
 
 | | Phosphoribosylaminoimidazole carboxylase with fructose-6-phosphate bound to the central channel of the octameric protein structure. | | 分子名称: | CHLORIDE ION, FORMIC ACID, FRUCTOSE -6-PHOSPHATE, ... | | 著者 | Filippova, E.V, Wawrzak, Z, Kudritska, M, Edwards, A, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2010-09-01 | | 公開日 | 2010-11-17 | | 最終更新日 | 2024-11-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Phosphoribosylaminoimidazole carboxylase with fructose-6-phosphate bound to the central channel of the octameric protein structure.
To be Published
|
|
6MLI
 
 | | Crystal structure of the periplasmic Lysine-, Arginine-, Ornithine-binding protein (LAO) R77A mutant from Salmonella typhimurium complexed with histidine | | 分子名称: | ACETATE ION, GLYCEROL, HISTIDINE, ... | | 著者 | Romero-Romero, S, Vergara, R, Espinoza-Perez, G, Rodriguez-Romero, A. | | 登録日 | 2018-09-27 | | 公開日 | 2019-08-07 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (1.883 Å) | | 主引用文献 | The interplay of protein-ligand and water-mediated interactions shape affinity and selectivity in the LAO binding protein.
Febs J., 287, 2020
|
|
6PA9
 
 | |
6MKX
 
 | | Crystal structure of the periplasmic Lysine-, Arginine-, Ornithine-binding protein (LAO) R77A mutant from Salmonella typhimurium | | 分子名称: | Lysine/arginine/ornithine-binding periplasmic protein | | 著者 | Romero-Romero, S, Vergara, R, Espinoza-Perez, G, Rodriguez-Romero, A. | | 登録日 | 2018-09-26 | | 公開日 | 2019-08-07 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.284 Å) | | 主引用文献 | The interplay of protein-ligand and water-mediated interactions shape affinity and selectivity in the LAO binding protein.
Febs J., 287, 2020
|
|
6MLE
 
 | | Crystal structure of the periplasmic Lysine-, Arginine-, Ornithine-binding protein (LAO) from Salmonella typhimurium complexed with arginine | | 分子名称: | ACETATE ION, ARGININE, Lysine/arginine/ornithine-binding periplasmic protein | | 著者 | Romero-Romero, S, Vergara, R, Espinoza-Perez, G, Rodriguez-Romero, A. | | 登録日 | 2018-09-27 | | 公開日 | 2019-08-07 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (1.858 Å) | | 主引用文献 | The interplay of protein-ligand and water-mediated interactions shape affinity and selectivity in the LAO binding protein.
Febs J., 287, 2020
|
|
3OOW
 
 | | Octameric structure of the phosphoribosylaminoimidazole carboxylase catalytic subunit from Francisella tularensis subsp. tularensis SCHU S4. | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, FORMIC ACID, ... | | 著者 | Filippova, E.V, Wawrzak, Z, Kudritska, M, Edwards, A, Savchenko, A, Anderson, F.W, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2010-08-31 | | 公開日 | 2010-09-15 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Octameric structure of the phosphoribosylaminoimidazole carboxylase catalytic subunit from Francisella tularensis subsp. tularensis SCHU S4.
To be Published
|
|
