1BWG
 
 | | DNA TRIPLEX WITH 5' AND 3' JUNCTIONS, NMR, 10 STRUCTURES | | 分子名称: | DNA (5'-D(*CP*TP*CP*TP*CP*T)-3'), DNA (5'-D(*GP*AP*CP*TP*GP*AP*GP*AP*GP*AP*CP*GP*TP*A)-3'), DNA (5'-D(*TP*AP*CP*GP*TP*CP*TP*CP*TP*CP*AP*GP*TP*C)-3') | | 著者 | Asensio, J.L, Brown, T, Lane, A.N. | | 登録日 | 1998-09-22 | | 公開日 | 1999-03-23 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution conformation of a parallel DNA triple helix with 5' and 3' triplex-duplex junctions.
Structure Fold.Des., 7, 1999
|
|
3K7C
 
 | |
6UVR
 
 | |
6GRC
 
 | | eukaryotic junction-resolving enzyme GEN-1 binding with Sodium | | 分子名称: | DNA (5'-D(*TP*AP*CP*CP*CP*AP*CP*CP*AP*CP*CP*GP*CP*TP*CP*A)-3'), DNA (5'-D(*TP*GP*AP*GP*CP*GP*GP*TP*GP*GP*TP*TP*GP*GP*T)-3'), MAGNESIUM ION, ... | | 著者 | Lilley, D.M.J, Liu, Y, Freeman, D.J. | | 登録日 | 2018-06-11 | | 公開日 | 2018-09-26 | | 最終更新日 | 2019-02-13 | | 実験手法 | X-RAY DIFFRACTION (2.452 Å) | | 主引用文献 | A monovalent ion in the DNA binding interface of the eukaryotic junction-resolving enzyme GEN1.
Nucleic Acids Res., 46, 2018
|
|
6GRD
 
 | | eukaryotic junction-resolving enzyme GEN-1 binding with Cesium | | 分子名称: | CESIUM ION, DNA (5'-D(*TP*AP*CP*CP*CP*AP*CP*CP*AP*CP*CP*GP*CP*TP*CP*A)-3'), DNA (5'-D(*TP*GP*AP*GP*CP*GP*GP*TP*GP*GP*TP*TP*GP*GP*T)-3'), ... | | 著者 | Lilley, D.M.J, Liu, Y, Freeman, D.J. | | 登録日 | 2018-06-11 | | 公開日 | 2018-09-26 | | 最終更新日 | 2018-11-28 | | 実験手法 | X-RAY DIFFRACTION (2.66 Å) | | 主引用文献 | A monovalent ion in the DNA binding interface of the eukaryotic junction-resolving enzyme GEN1.
Nucleic Acids Res., 46, 2018
|
|
5YWW
 
 | | Archael RuvB-like Holiday junction helicase | | 分子名称: | GLYCEROL, Nucleotide binding protein PINc | | 著者 | Zhai, B, Yuan, Z, Han, X, DuPrez, K, Shen, Y, Fan, L. | | 登録日 | 2017-11-30 | | 公開日 | 2018-06-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.33 Å) | | 主引用文献 | The archaeal ATPase PINA interacts with the helicase Hjm via its carboxyl terminal KH domain remodeling and processing replication fork and Holliday junction.
Nucleic Acids Res., 46, 2018
|
|
3UDO
 
 | | Crystal structure of putative isopropylamlate dehydrogenase from Campylobacter jejuni | | 分子名称: | 1,2-ETHANEDIOL, 3-isopropylmalate dehydrogenase, SULFATE ION | | 著者 | Tkaczuk, K.L, Chruszcz, M, Blus, B.J, Onopriyenko, O, Grimshaw, S, Savchenko, A, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2011-10-28 | | 公開日 | 2011-11-09 | | 最終更新日 | 2022-04-13 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structure of putative isopropylamlate dehydrogenase from Campylobacter jejuni
To be Published
|
|
5F4H
 
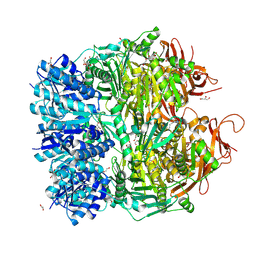 | | Archael RuvB-like Holiday junction helicase | | 分子名称: | 1,2-ETHANEDIOL, GLYCEROL, Nucleotide binding protein PINc | | 著者 | Zhai, B, DuPrez, K.T, Doukov, T.I, Shen, Y, Fan, L. | | 登録日 | 2015-12-03 | | 公開日 | 2016-12-21 | | 最終更新日 | 2017-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.699 Å) | | 主引用文献 | Structure and Function of a Novel ATPase that Interacts with Holliday Junction Resolvase Hjc and Promotes Branch Migration.
J. Mol. Biol., 429, 2017
|
|
6VE7
 
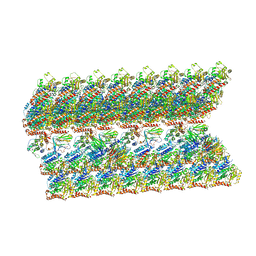 | | The inner junction complex of Chlamydomonas reinhardtii doublet microtubule | | 分子名称: | Cilia- and flagella-associated protein 20, FAP276, FAP52, ... | | 著者 | Khalifa, A.A.Z, Ichikawa, M, Bui, K.H. | | 登録日 | 2019-12-30 | | 公開日 | 2020-02-05 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | The inner junction complex of the cilia is an interaction hub that involves tubulin post-translational modifications.
Elife, 9, 2020
|
|
5T0O
 
 | |
6JN8
 
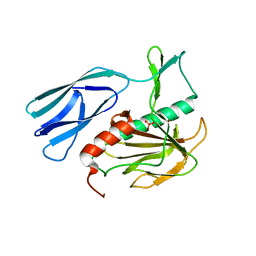 | | Structure of H216A mutant open form peptidoglycan peptidase | | 分子名称: | Peptidase M23, SULFATE ION, ZINC ION | | 著者 | Min, K.J, An, D.R, Yoon, H.J, Suh, S.W, Lee, H.H. | | 登録日 | 2019-03-13 | | 公開日 | 2020-01-15 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.106 Å) | | 主引用文献 | Peptidoglycan reshaping by a noncanonical peptidase for helical cell shape in Campylobacter jejuni.
Nat Commun, 11, 2020
|
|
1XNS
 
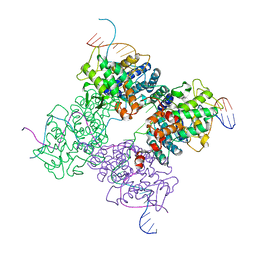 | | Peptide trapped Holliday junction intermediate in Cre-loxP recombination | | 分子名称: | Recombinase CRE, loxP DNA | | 著者 | Ghosh, K, Lau, C.K, Guo, F, Segall, A.M, Van Duyne, G.D. | | 登録日 | 2004-10-05 | | 公開日 | 2004-12-14 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Peptide trapping of the Holliday junction intermediate in Cre-loxP site-specific recombination.
J.Biol.Chem., 280, 2005
|
|
6VO6
 
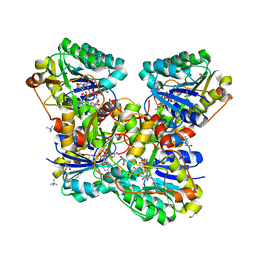 | | Crystal Structure of Cj1427, an Essential NAD-dependent Dehydrogenase from Campylobacter jejuni, in the Presence of NADH and GDP | | 分子名称: | 1,2-ETHANEDIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, CHLORIDE ION, ... | | 著者 | Anderson, T.K, Spencer, K.D, Thoden, J.B, Huddleston, J.P, Raushel, F.M, Holden, H.M. | | 登録日 | 2020-01-30 | | 公開日 | 2020-04-01 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structural Analysis of Cj1427, an Essential NAD-Dependent Dehydrogenase for the Biosynthesis of the Heptose Residues in the Capsular Polysaccharides ofCampylobacter jejuni.
Biochemistry, 59, 2020
|
|
6VO8
 
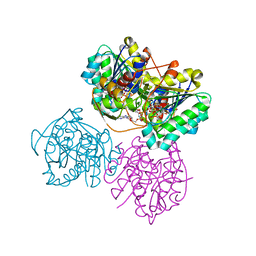 | | X-ray structure of the Cj1427 in the presence of NADH and GDP-D-glycero-D-mannoheptose, an essential NAD-dependent dehydrogenase from Campylobacter jejuni | | 分子名称: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Putative sugar-nucleotide epimerase/dehydratease, [[(2~{R},3~{S},4~{R},5~{R})-5-(2-azanyl-6-oxidanylidene-3~{H}-purin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{S},5~{S},6~{S})-6-[(1~{S})-1,2-bis(oxidanyl)ethyl]-3,4,5-tris(oxidanyl)oxan-2-yl] hydrogen phosphate | | 著者 | Spencer, K.D, Anderson, T.K, Thoden, J.B, Huddleston, J.P, Raushel, F.M, Holden, H.M. | | 登録日 | 2020-01-30 | | 公開日 | 2020-03-25 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural Analysis of Cj1427, an Essential NAD-Dependent Dehydrogenase for the Biosynthesis of the Heptose Residues in the Capsular Polysaccharides ofCampylobacter jejuni.
Biochemistry, 59, 2020
|
|
6JMZ
 
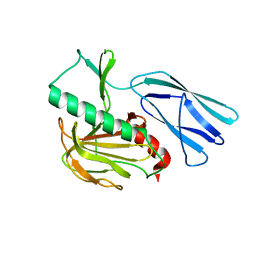 | | Structure of H247A mutant open form peptidoglycan peptidase | | 分子名称: | Peptidase M23, ZINC ION | | 著者 | Min, K.J, An, D.R, Yoon, H.J, Suh, S.W, Lee, H.H. | | 登録日 | 2019-03-13 | | 公開日 | 2020-01-15 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | Peptidoglycan reshaping by a noncanonical peptidase for helical cell shape in Campylobacter jejuni.
Nat Commun, 11, 2020
|
|
6JMX
 
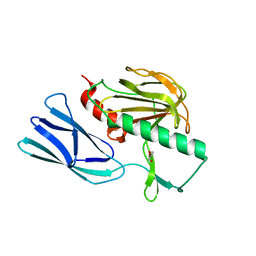 | | Structure of open form of peptidoglycan peptidase | | 分子名称: | D(-)-TARTARIC ACID, GLYCEROL, Peptidase M23, ... | | 著者 | Min, K.J, An, D.R, Yoon, H.J, Suh, S.W, Lee, H.H. | | 登録日 | 2019-03-13 | | 公開日 | 2020-01-15 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.859 Å) | | 主引用文献 | Peptidoglycan reshaping by a noncanonical peptidase for helical cell shape in Campylobacter jejuni.
Nat Commun, 11, 2020
|
|
6JN1
 
 | | Structure of H247A mutant peptidoglycan peptidase complex with penta peptide | | 分子名称: | C0O-DAL-DAL, Peptidase M23, ZINC ION | | 著者 | Min, K.J, An, D.R, Yoon, H.J, Suh, S.W, Lee, H.H. | | 登録日 | 2019-03-13 | | 公開日 | 2020-01-15 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.382 Å) | | 主引用文献 | Peptidoglycan reshaping by a noncanonical peptidase for helical cell shape in Campylobacter jejuni.
Nat Commun, 11, 2020
|
|
196D
 
 | |
6JN0
 
 | | Structure of H247A mutant peptidoglycan peptidase complex with tetra-tri peptide | | 分子名称: | C0O-DAL-API, Peptidase M23, ZINC ION | | 著者 | Min, K.J, An, D.R, Yoon, H.J, Suh, S.W, Lee, H.H. | | 登録日 | 2019-03-13 | | 公開日 | 2020-01-15 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.164 Å) | | 主引用文献 | Peptidoglycan reshaping by a noncanonical peptidase for helical cell shape in Campylobacter jejuni.
Nat Commun, 11, 2020
|
|
6JN7
 
 | | Structure of H216A mutant closed form peptidoglycan peptidase | | 分子名称: | D(-)-TARTARIC ACID, Peptidase M23, ZINC ION | | 著者 | Min, K.J, An, D.R, Yoon, H.J, Suh, S.W, Lee, H.H. | | 登録日 | 2019-03-13 | | 公開日 | 2020-01-15 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.04 Å) | | 主引用文献 | Peptidoglycan reshaping by a noncanonical peptidase for helical cell shape in Campylobacter jejuni.
Nat Commun, 11, 2020
|
|
6JMY
 
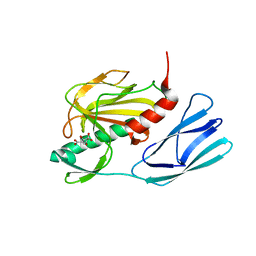 | | Structure of wild type closed form of peptidoglycan peptidase | | 分子名称: | CITRIC ACID, Peptidase M23, ZINC ION | | 著者 | Min, K.J, An, D.R, Yoon, H.J, Suh, S.W, Lee, H.H. | | 登録日 | 2019-03-13 | | 公開日 | 2020-01-15 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.661 Å) | | 主引用文献 | Peptidoglycan reshaping by a noncanonical peptidase for helical cell shape in Campylobacter jejuni.
Nat Commun, 11, 2020
|
|
1XO0
 
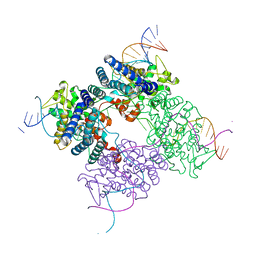 | | High resolution structure of the holliday junction intermediate in cre-loxp site-specific recombination | | 分子名称: | Recombinase CRE, loxP | | 著者 | Ghosh, K, Lau, C.K, Guo, F, Segall, A.M, Van Duyne, G.D. | | 登録日 | 2004-10-05 | | 公開日 | 2004-12-14 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Peptide trapping of the Holliday junction intermediate in Cre-loxP site-specific recombination.
J.Biol.Chem., 280, 2005
|
|
2CRX
 
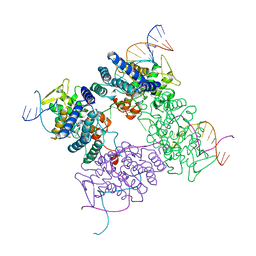 | |
7C7Z
 
 | |
7JHV
 
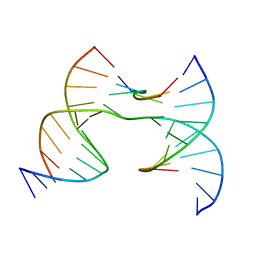 | | Self-assembly of a 3D DNA crystal lattice (4x6 junction version) containing the J36 immobile Holliday junction with R3 symmetry | | 分子名称: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*GP*TP*GP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*CP*GP*TP*CP*TP*GP*C)-3'), DNA (5'-D(P*CP*GP*AP*CP*AP*CP*TP*CP*A)-3'), ... | | 著者 | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | 登録日 | 2020-07-21 | | 公開日 | 2021-07-14 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.058 Å) | | 主引用文献 | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
