1K4H
 
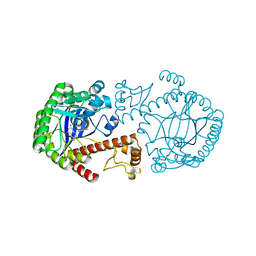 | | CRYSTAL STRUCTURE OF TRNA-GUANINE TRANSGLYCOSYLASE (TGT) COMPLEXED WITH 2,6-Diamino-8-propylsulfanylmethyl-3H-quinazoline-4-one | | 分子名称: | 2,6-DIAMINO-8-PROPYLSULFANYLMETHYL-3H-QUINAZOLINE-4-ONE, TRNA-GUANINE-TRANSGLYCOSYLASE, ZINC ION | | 著者 | Brenk, R, Meyer, E.A, Castellano, R.K, Furler, M, Stubbs, M.T, Klebe, G, Diederich, F. | | 登録日 | 2001-10-08 | | 公開日 | 2002-04-24 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | De novo design, synthesis, and in vitro evaluation of inhibitors for prokaryotic tRNA-guanine transglycosylase: a dramatic sulfur effect on binding affinity.
Chembiochem, 3, 2002
|
|
1FSV
 
 | |
1FSD
 
 | |
1FFQ
 
 | | CRYSTAL STRUCTURE OF CHITINASE A COMPLEXED WITH ALLOSAMIDIN | | 分子名称: | 2-acetamido-2-deoxy-beta-D-allopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-allopyranose, ALLOSAMIZOLINE, CHITINASE A | | 著者 | Papanikolau, Y, Tavlas, G, Vorgias, C.E, Petratos, K. | | 登録日 | 2000-07-26 | | 公開日 | 2003-02-11 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | De novo purification scheme and crystallization conditions yield high-resolution structures of chitinase A and its complex with the inhibitor allosamidin.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1K4G
 
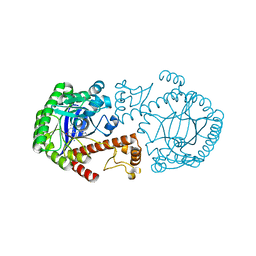 | | CRYSTAL STRUCTURE OF TRNA-GUANINE TRANSGLYCOSYLASE (TGT) COMPLEXED WITH 2,6-DIAMINO-8-(1H-IMIDAZOL-2-YLSULFANYLMETHYL)-3H-QUINAZOLINE-4-ONE | | 分子名称: | 2,6-DIAMINO-8-(1H-IMIDAZOL-2-YLSULFANYLMETHYL)-3H-QUINAZOLINE-4-ONE, TRNA-GUANINE TRANSGLYCOSYLASE, ZINC ION | | 著者 | Brenk, R, Meyer, E.A, Castellano, R.K, Furler, M, Stubbs, M.T, Klebe, G, Diederich, F. | | 登録日 | 2001-10-08 | | 公開日 | 2002-04-24 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | De novo design, synthesis, and in vitro evaluation of inhibitors for prokaryotic tRNA-guanine transglycosylase: a dramatic sulfur effect on binding affinity.
ChemBioChem, 3, 2002
|
|
6L0L
 
 | |
2BE9
 
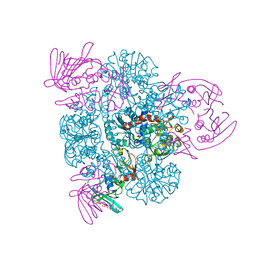 | | Crystal structure of the CTP-liganded (T-State) aspartate transcarbamoylase from the extremely thermophilic archaeon Sulfolobus acidocaldarius | | 分子名称: | Aspartate carbamoyltransferase, Aspartate carbamoyltransferase regulatory chain, CYTIDINE-5'-TRIPHOSPHATE, ... | | 著者 | De Vos, D, Savvides, S.N, Van Beeumen, J.J. | | 登録日 | 2005-10-23 | | 公開日 | 2006-10-31 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Crystal structure of Sulfolobus acidocaldarius aspartate carbamoyltransferase in complex with its allosteric activator CTP.
Biochem.Biophys.Res.Commun., 372, 2008
|
|
2BE7
 
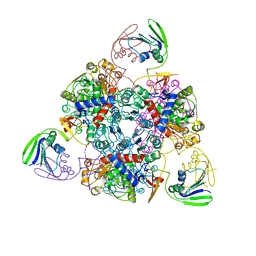 | | Crystal structure of the unliganded (T-state) aspartate transcarbamoylase of the psychrophilic bacterium Moritella profunda | | 分子名称: | Aspartate Carbamoyltransferase Catalytic Chain, Aspartate Carbamoyltransferase Regulatory Chain, SULFATE ION, ... | | 著者 | De Vos, D, Savvides, S.N, Van Beeumen, J. | | 登録日 | 2005-10-23 | | 公開日 | 2006-10-31 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | Structural investigation of cold activity and regulation of aspartate carbamoyltransferase from the extreme psychrophilic bacterium Moritella profunda.
J.Mol.Biol., 365, 2007
|
|
3FBX
 
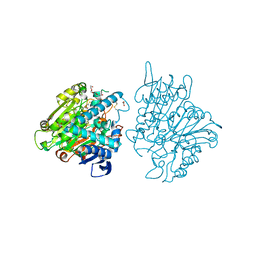 | | Crystal structure of the lysosomal 66.3 kDa protein from mouse solved by S-SAD | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | 著者 | Lakomek, K, Dickmanns, A, Mueller, U, Ficner, R. | | 登録日 | 2008-11-20 | | 公開日 | 2009-03-03 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | De novo sulfur SAD phasing of the lysosomal 66.3 kDa protein from mouse
Acta Crystallogr.,Sect.D, 65, 2009
|
|
5L31
 
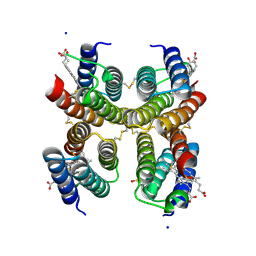 | |
5L32
 
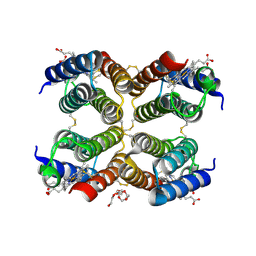 | |
3B5L
 
 | |
1EDQ
 
 | |
1EKY
 
 | |
7RKC
 
 | | Computationally designed tunable C2 symmetric tandem repeat homodimer, D_3_633 | | 分子名称: | ACETATE ION, D_3_633 | | 著者 | Kennedy, M.A, Stoddard, B.L, Hicks, D.R, Bera, A.K. | | 登録日 | 2021-07-22 | | 公開日 | 2022-05-18 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | De novo design of protein homodimers containing tunable symmetric protein pockets.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
3HOJ
 
 | |
1Q7O
 
 | | Determination of f-MLF-OH Peptide Structure with solid-state magic-angle spinning NMR Spectroscopy | | 分子名称: | chemotactic peptide | | 著者 | Rienstra, C.M, Tucker-Kellogg, L, Jaroniec, C.P, Hohwy, M, Reif, B, McMahon, M.T, Tidor, B, Lozano-Perez, T, Griffin, R.G. | | 登録日 | 2003-08-19 | | 公開日 | 2003-09-09 | | 最終更新日 | 2022-03-02 | | 実験手法 | SOLID-STATE NMR | | 主引用文献 | De novo determination of peptide structure with solid-state magic-angle spinning NMR Spectroscopy
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1PSV
 
 | |
2MUZ
 
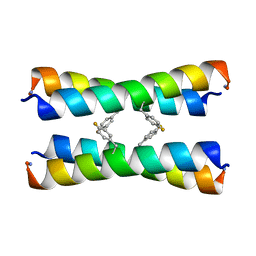 | | ssNMR structure of a designed rocker protein | | 分子名称: | designed rocker protein | | 著者 | Wang, T, Joh, N, Wu, Y, DeGrado, W.F, Hong, M. | | 登録日 | 2014-09-18 | | 公開日 | 2014-12-24 | | 最終更新日 | 2015-01-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | De novo design of a transmembrane Zn2+-transporting four-helix bundle.
Science, 346, 2014
|
|
7QDI
 
 | | Structure of octameric left-handed 310-helix bundle: D-310HD | | 分子名称: | 1,2-ETHANEDIOL, 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 2-(2-METHOXYETHOXY)ETHANOL, ... | | 著者 | Kumar, P, Paterson, N.G, Woolfson, D.N. | | 登録日 | 2021-11-27 | | 公開日 | 2022-04-27 | | 最終更新日 | 2022-07-27 | | 実験手法 | X-RAY DIFFRACTION (2.34 Å) | | 主引用文献 | De novo design of discrete, stable 3 10 -helix peptide assemblies.
Nature, 607, 2022
|
|
7QDJ
 
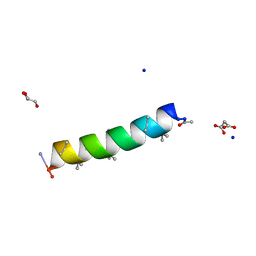 | | Racemic structure of PK-10 and PK-11 | | 分子名称: | GLYCEROL, MALONATE ION, PK-10+PK-11, ... | | 著者 | Kumar, P, Paterson, N.G, Woolfson, D.N. | | 登録日 | 2021-11-27 | | 公開日 | 2022-04-27 | | 最終更新日 | 2022-07-27 | | 実験手法 | X-RAY DIFFRACTION (1.44 Å) | | 主引用文献 | De novo design of discrete, stable 3 10 -helix peptide assemblies.
Nature, 607, 2022
|
|
7UDK
 
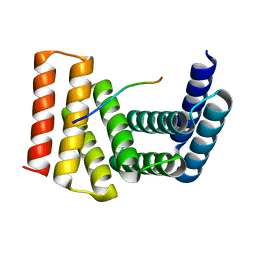 | | Crystal structure of designed helical repeat protein RPB_LRP2_R4 bound to LRPx4 peptide | | 分子名称: | 4xLRP, Designed helical repeat protein (DHR) RPB_LRP2_R4 | | 著者 | Chang, Y, Redler, R.L, Bhabha, G, Ekiert, D.C. | | 登録日 | 2022-03-20 | | 公開日 | 2023-03-22 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (3.18 Å) | | 主引用文献 | De novo design of modular peptide-binding proteins by superhelical matching.
Nature, 616, 2023
|
|
7UDL
 
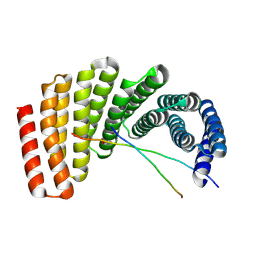 | | Crystal structure of designed helical repeat protein RPB_PLP1_R6 bound to PLPx6 peptide | | 分子名称: | 1,2-ETHANEDIOL, 6xPLP Peptide, Designed helical repeat protein (DHR) RPB_PLP1_R6 | | 著者 | Chang, Y, Redler, R.L, Bhabha, G, Ekiert, D.C. | | 登録日 | 2022-03-20 | | 公開日 | 2023-03-22 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | De novo design of modular peptide-binding proteins by superhelical matching.
Nature, 616, 2023
|
|
7UDN
 
 | |
7VU4
 
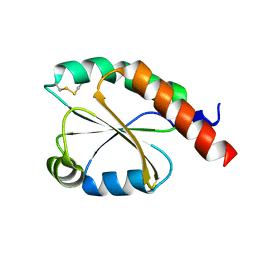 | | de novo design based on 1r26 | | 分子名称: | de novo design protein | | 著者 | Zhang, L. | | 登録日 | 2021-11-01 | | 公開日 | 2022-06-08 | | 最終更新日 | 2023-06-28 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Rotamer-free protein sequence design based on deep learning and self-consistency.
Nat Comput Sci, 2023
|
|
